Optical Monitoring of Breathing Patterns and Tissue Oxygenation: A Potential Application in COVID-19 Screening and Monitoring
Abstract
1. Introduction
Objective
2. Methods
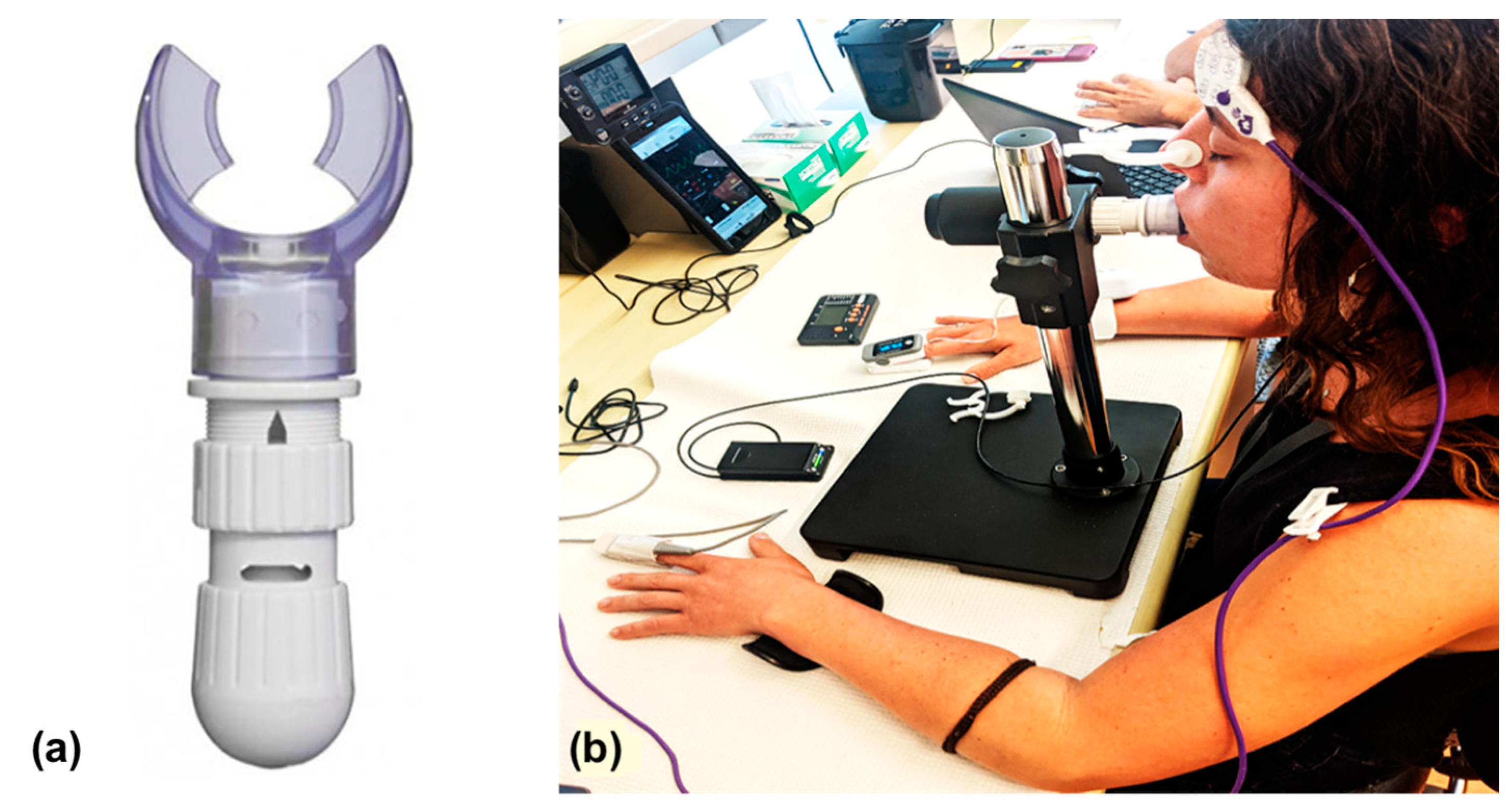
2.1. Instrumentation
2.2. Participants
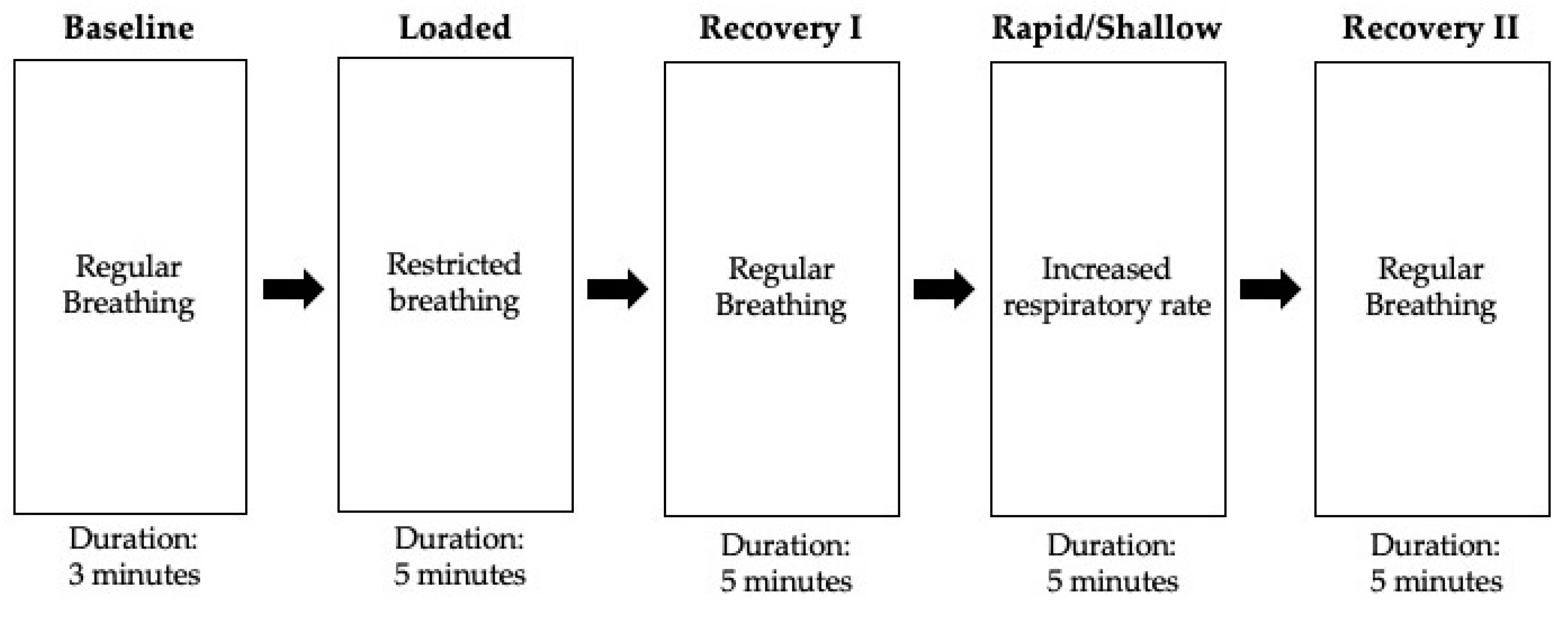
2.3. Experimental Protocol
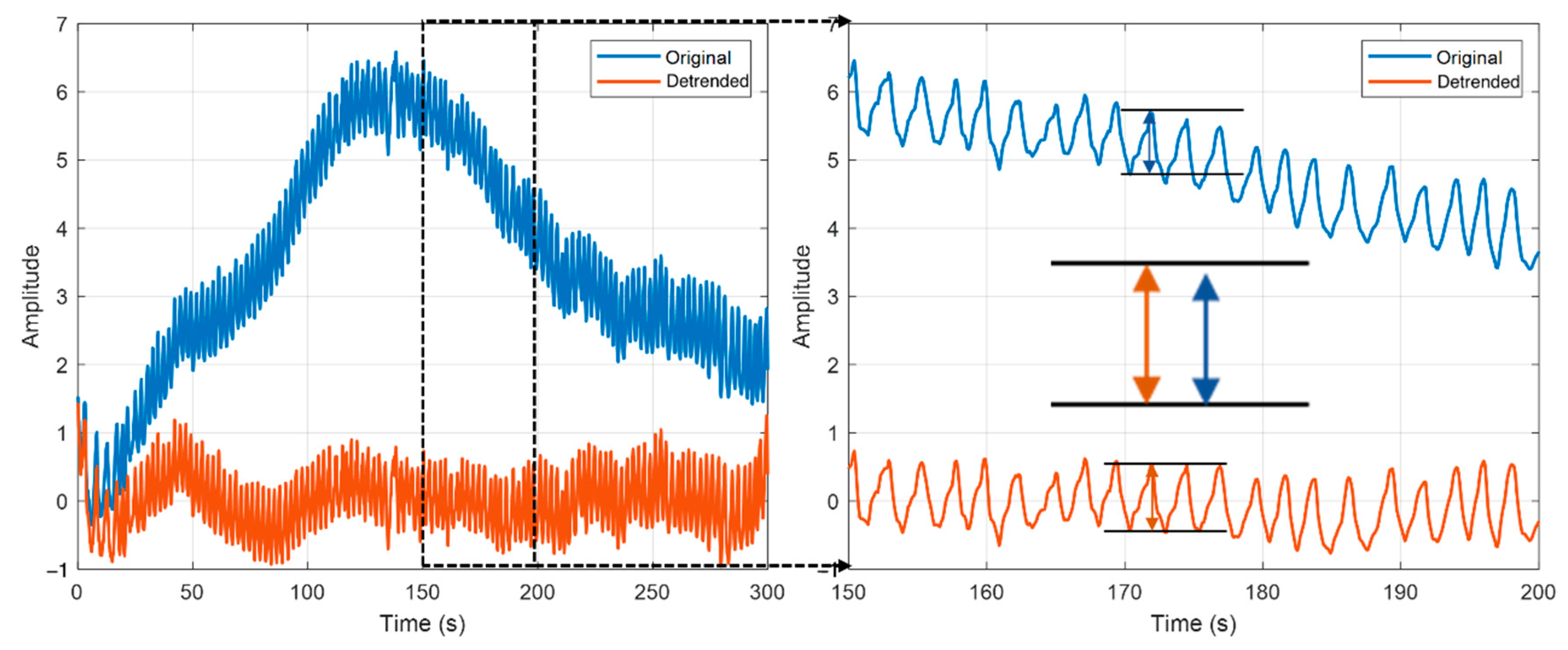
2.4. Data Pre-Processing
2.5. Data Analysis
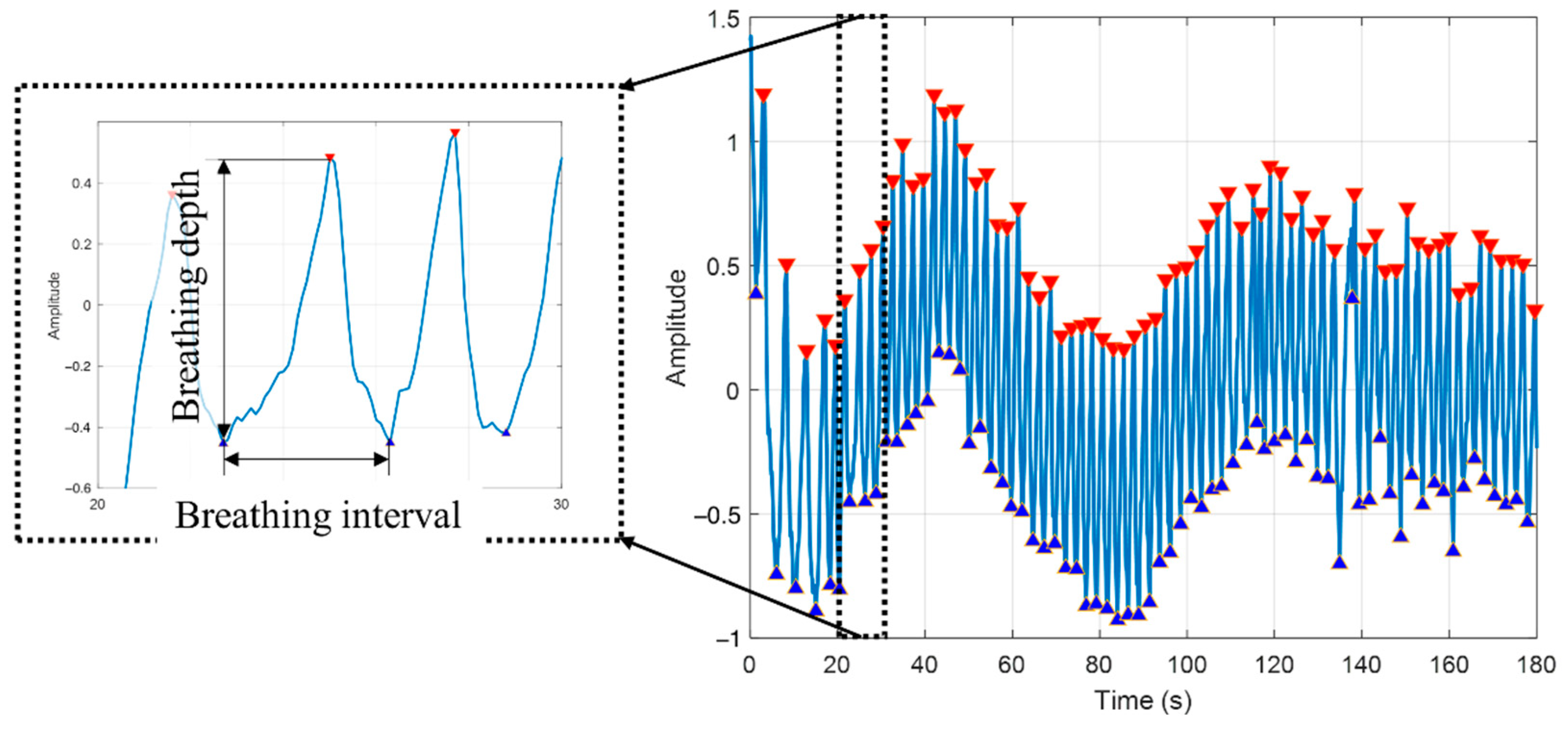
2.5.1. Peak Detection
2.5.2. Feature Selection
2.5.3. Machine Learning
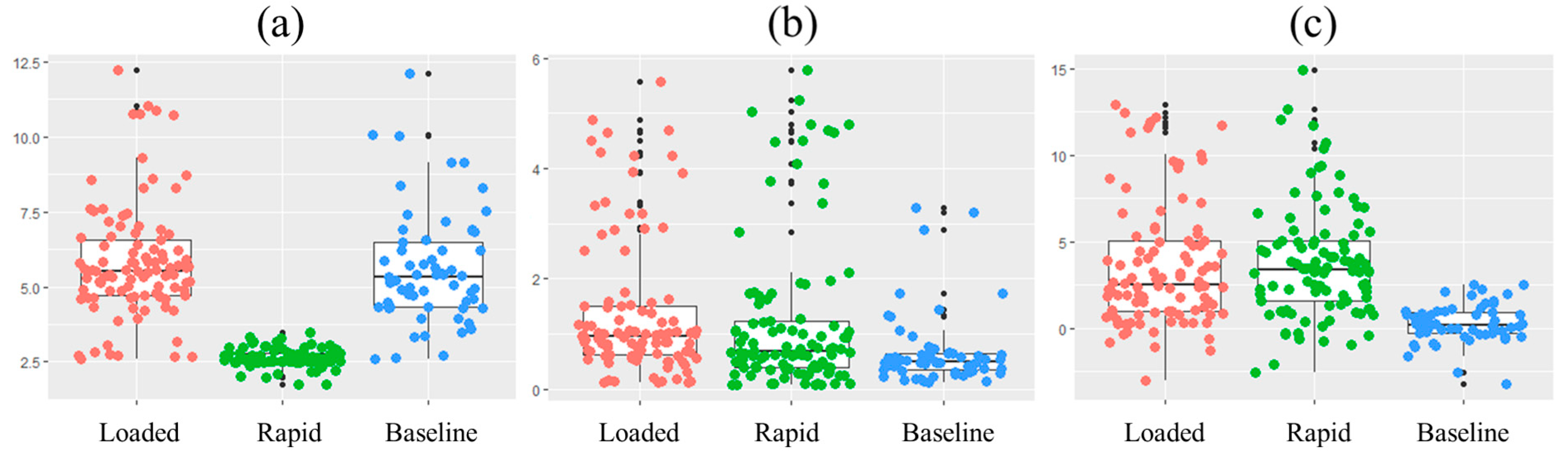
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hashmi, H.A.S.; Asif, H.M. Early Detection and Assessment of COVID-19. Front. Med. 2020, 7, 311. [Google Scholar] [CrossRef] [PubMed]
- Horgan, C.; Goody, B.; Knapp, D.; Fitterman, L. The role of mail service pharmacies. Health Aff. 1990, 9, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Ardebili, A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020, 20, 453–454. [Google Scholar] [CrossRef] [PubMed]
- Peeling, R.W.; Olliaro, P.L.; Boeras, D.I.; Fongwen, N. Scaling up COVID-19 rapid antigen tests: Promises and challenges. Lancet Infect. Dis. 2021, 21, e290–e295. [Google Scholar] [CrossRef]
- Cajanding, R. Silent Hypoxia in COVID-19 Pneumonia: State of Knowledge, Pathophysiology, Mechanisms, and Management. AACN Adv. Crit. Care 2022, 33, 143–153. [Google Scholar] [CrossRef]
- Luks, A.M.; Swenson, E.R. Pulse oximetry for monitoring patients with COVID-19 at home potential pitfalls and practical guidance. Ann. Am. Thorac. Soc. 2020, 17, 1040–1046. [Google Scholar] [CrossRef]
- Mo, X.; Jian, W.; Su, Z.; Chen, M.; Peng, H.; Peng, P.; Lei, C.; Chen, R.; Zhong, N.; Li, S. Abnormal pulmonary function in COVID-19 patients at time of hospital discharge. Eur. Respir. J. 2020, 55, 2001217. [Google Scholar] [CrossRef]
- Goonasekera, C.; Peiris, P.; Oswald, L.; Sheikh, A. SpO2: How Low is Too Low? J. Med. Physiol. Ther. 2017, 1, 105. [Google Scholar]
- Louw, A.; Cracco, C.; Cerf, C.; Harf, A.; Duvaldestin, P.; Lemaire, F.; Brochard, L. Accuracy of pulse oximetry in the intensive care unit. Intensive Care Med. 2001, 27, 1606–1613. [Google Scholar] [CrossRef]
- Tobias, J.D. Cerebral oximetry monitoring with near infrared spectroscopy detects alterations in oxygenation before pulse oximetry. J. Intensive Care Med. 2008, 23, 384–388. [Google Scholar] [CrossRef]
- Trivedi, N.S.; Ghouri, A.F.; Shah, N.K.; Lai, E.; Barker, S.J. Effects of motion, ambient light, and hypoperfusion on pulse oximeter function. J. Clin. Anesth. 1997, 9, 179–183. [Google Scholar] [CrossRef]
- Philip, K.E.J.; Bennett, B.; Fuller, S.; Lonergan, B.; McFadyen, C.; Burns, J.; Tidswell, R.; Vlachou, A. Working accuracy of pulse oximetry in COVID-19 patients stepping down from intensive care: A clinical evaluation. BMJ Open Respir. Res. 2020, 7, e000778. [Google Scholar] [CrossRef] [PubMed]
- Park, S.B.; Khattar, D. Tachypnea. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Izcovich, A.; Ragusa, M.A.; Tortosa, F.; Lavena Marzio, M.A.; Agnoletti, C.; Bengolea, A.; Ceirano, A.; Espinosa, F.; Saavedra, E.; Sanguine, V.; et al. Prognostic factors for severity and mortality in patients infected with COVID-19: A systematic review. PLoS ONE 2020, 15, e0241955. [Google Scholar] [CrossRef] [PubMed]
- Pagliano, P.; Sellitto, C.; Conti, V.; Ascione, T.; Esposito, S. Characteristics of viral pneumonia in the COVID-19 era: An update. Infection 2021, 49, 607–616. [Google Scholar] [CrossRef]
- Higgins, V.; Sohaei, D.; Diamandis, E.P.; Prassas, I. COVID-19: From an acute to chronic disease? Potential long-term health consequences. Crit. Rev. Clin. Lab. Sci. 2021, 58, 297–310. [Google Scholar] [CrossRef]
- Raghu, G.; Wilson, K.C. COVID-19 interstitial pneumonia: Monitoring the clinical course in survivors. Lancet Respir. Med. 2020, 8, 839–842. [Google Scholar] [CrossRef]
- Orzes, N.; Pini, L.; Levi, G.; Uccelli, S.; Cettolo, F.; Tantucci, C. A prospective evaluation of lung function at three and six months in patients with previous SARS-CoV-2 pneumonia. Respir. Med. 2020, 186, 106541. [Google Scholar] [CrossRef]
- Wu, Q.; Zhong, L.; Li, H.; Guo, J.; Li, Y.; Hou, X.; Yang, F.; Xie, Y.; Li, L.; Xing, Z. A Follow-Up Study of Lung Function and Chest Computed Tomography at 6 Months after Discharge in Patients with Coronavirus Disease 2019. Can. Respir. J. 2021, 2021, 6692409. [Google Scholar] [CrossRef] [PubMed]
- Shadgan, B.; Reid, W.D.; Gharakhanlou, R.; Stothers, L.; MacNab, A.J. Wireless near-infrared spectroscopy of skeletal muscle oxygenation and hemodynamics during exercise and ischemia. Spectroscopy 2009, 23, 233–241. [Google Scholar] [CrossRef]
- Hagino, I.; Anttila, V.; Zurakowski, D.; Duebener, L.F.; Lidov, H.G.; Jonas, R.A. Tissue oxygenation index is a useful monitor of histologic and neurologic outcome after cardiopulmonary bypass in piglets. J. Thorac. Cardiovasc. Surg. 2005, 130, 384–392. [Google Scholar] [CrossRef]
- Murkin, J.M.; Arango, M. Near-infrared spectroscopy as an index of brain and tissue oxygenation. Br. J. Anaesth. 2009, 103 (Suppl. 1), i3–i13. [Google Scholar] [CrossRef] [PubMed]
- Macnab, A.J.; Shadgan, B.; Stothers, L. Review: The evolution of wireless near infrared spectroscopy applications in urology and rationale for clinical use. J. Near Infrared Spectrosc. 2012, 20, 57–73. [Google Scholar] [CrossRef]
- Gagnon, R.E.; Macnab, A.J. Near infrared spectroscopy (NIRS) in the clinical setting—An adjunct to monitoring during diagnosis and treatment. Spectroscopy 2005, 19, 221–233. [Google Scholar] [CrossRef]
- Reid, W.D.; Sheel, A.W.; Shadgan, B.; Garland, S.J.; Road, J.D. Recruitment and Deoxygenation of Selected Respiratory and Skeletal Muscles During Incremental Loading in Stable COPD Patients. J. Cardiopulm. Rehabil. Prev. 2016, 36, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Basoudan, N.; Shadgan, B.; Guenette, J.A.; Road, J.; Reid, W.D. Effect of acute hypoxia on inspiratory muscle oxygenation during incremental inspiratory loading in healthy adults. Eur. J. Appl. Physiol. 2016, 116, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Shadgan, B.; Guenette, J.A.; Sheel, A.W.; Reid, W.D. Sternocleidomastoid muscle deoxygenation in response to incremental inspiratory threshold loading measured by near infrared spectroscopy. Respir. Physiol. Neurobiol. 2011, 178, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, J.H.; McDonald, G.; Alton, H.; Gordon, S.B. Pneumonia in the immunocompetent patient. Br. J. Radiol. 2010, 83, 998–1009. [Google Scholar] [CrossRef]
- Morettini, E.; Turchini, F.; Tofani, L.; Villa, G.; Ricci, Z.; Romagnoli, S. Intraoperative core temperature monitoring: Accuracy and precision of zero-heat flux heated controlled servo sensor compared with esophageal temperature during major surgery; the ESOSPOT study. J. Clin. Monit. Comput. 2020, 34, 1111–1119. [Google Scholar] [CrossRef]
- Ho, T.K. Random decision forests. In Proceedings of the Third International Conference on Document Analysis and Recognition (ICDAR ’95), Montreal, QC, Canada, 14–16 August 1995; Volume 1, p. 278. [Google Scholar]
- Rothan, H.A.; Byrareddy, S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020, 109, 102433. [Google Scholar] [CrossRef]
- Lei, J.; Li, J.; Li, X.; Qi, X. CT Imaging of the 2019 Novel Coronavirus (2019-nCoV) Pneumonia. Radiology 2020, 295, 18. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.; Lau, E.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lei, X.; Zhang, L.; Zhang, L.; Dong, W. The application of near-infrared spectroscopy in oxygen therapy for premature infants. J. Matern.-Fetal Neonatal Med. 2020, 33, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Brochard, L.; Martin, G.S.; Blanch, L.; Pelosi, P.; Belda, F.J.; Jubran, A.; Gattinoni, L.; Mancebo, J.; Ranieri, V.M.; Richard, J.C.; et al. Clinical review: Respiratory monitoring in the ICU—A consensus of 16. Crit. Care 2012, 16, 219. [Google Scholar] [CrossRef] [PubMed]
- Gargani, L.; Soliman-Aboumarie, H.; Volpicelli, G.; Corradi, F.; Pastore, M.C.; Cameli, M. Why, when, and how to use lung ultrasound during the COVID-19 pandemic: Enthusiasm and caution. Eur. Heart J. Cardiovasc. Imaging 2020, 21, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.S.; Read, S.W.; Fauci, A.S. Therapy for Early COVID-19: A Critical Need. JAMA 2020, 324, 2149–2150. [Google Scholar] [CrossRef] [PubMed]
- Bardakci, M.I.; Ozturk, E.N.; Ozkarafakili, M.A.; Ozkurt, H.; Yanc, U.; Yildiz Sevgi, D. Evaluation of long-term radiological findings, pulmonary functions, and health-related quality of life in survivors of severe COVID-19. J. Med. Virol. 2021, 93, 5574–5581. [Google Scholar] [CrossRef]
- Huang, Y.; Tan, C.; Wu, J.; Chen, M.; Wang, Z.; Luo, L.; Zhou, X.; Liu, X.; Huang, X.; Yuan, S.; et al. Impact of coronavirus disease 2019 on pulmonary function in early convalescence phase. Respir. Res. 2020, 21, 163. [Google Scholar] [CrossRef]
- Daniels, J.; Wadekar, S.; DeCubellis, K.; Jackson, G.W.; Chiu, A.S.; Pagneux, Q.; Saada, H.; Engelmann, I.; Ogiez, J.; Loze-Warot, D.; et al. A mask-based diagnostic platform for point-of-care screening of COVID-19. Biosens. Bioelectron. 2021, 192, 113486. [Google Scholar] [CrossRef]
- Giovannini, G.; Haick, H.; Garoli, D. Detecting COVID-19 from Breath: A Game Changer for a Big Challenge. ACS Sens. 2021, 6, 1408–1417. [Google Scholar] [CrossRef]
- Ravi, N.; Cortade, D.L.; Ng, E.; Wang, S.X. Diagnostics for SARS-CoV-2 detection: A comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020, 165, 112454. [Google Scholar] [CrossRef]
- Shan, B.; Broza, Y.Y.; Li, W.; Wang, Y.; Wu, S.; Liu, Z.; Wang, J.; Gui, S.; Wang, L.; Zhang, Z.; et al. Multiplexed Nanomaterial-Based Sensor Array for Detection of COVID-19 in Exhaled Breath. ACS Nano 2020, 14, 12125–12132. [Google Scholar] [CrossRef] [PubMed]


| Features | Weighted Accuracy | F1 Score |
|---|---|---|
| Depth, interval, O2Hb amplitude | 0.87 | 0.86 |
| Depth, interval | 0.79 | 0.77 |
| Interval, O2Hb amplitude | 0.78 | 0.76 |
| Depth, O2Hb amplitude | 0.63 | 0.62 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mah, A.J.; Nguyen, T.; Ghazi Zadeh, L.; Shadgan, A.; Khaksari, K.; Nourizadeh, M.; Zaidi, A.; Park, S.; Gandjbakhche, A.H.; Shadgan, B. Optical Monitoring of Breathing Patterns and Tissue Oxygenation: A Potential Application in COVID-19 Screening and Monitoring. Sensors 2022, 22, 7274. https://doi.org/10.3390/s22197274
Mah AJ, Nguyen T, Ghazi Zadeh L, Shadgan A, Khaksari K, Nourizadeh M, Zaidi A, Park S, Gandjbakhche AH, Shadgan B. Optical Monitoring of Breathing Patterns and Tissue Oxygenation: A Potential Application in COVID-19 Screening and Monitoring. Sensors. 2022; 22(19):7274. https://doi.org/10.3390/s22197274
Chicago/Turabian StyleMah, Aaron James, Thien Nguyen, Leili Ghazi Zadeh, Atrina Shadgan, Kosar Khaksari, Mehdi Nourizadeh, Ali Zaidi, Soongho Park, Amir H. Gandjbakhche, and Babak Shadgan. 2022. "Optical Monitoring of Breathing Patterns and Tissue Oxygenation: A Potential Application in COVID-19 Screening and Monitoring" Sensors 22, no. 19: 7274. https://doi.org/10.3390/s22197274
APA StyleMah, A. J., Nguyen, T., Ghazi Zadeh, L., Shadgan, A., Khaksari, K., Nourizadeh, M., Zaidi, A., Park, S., Gandjbakhche, A. H., & Shadgan, B. (2022). Optical Monitoring of Breathing Patterns and Tissue Oxygenation: A Potential Application in COVID-19 Screening and Monitoring. Sensors, 22(19), 7274. https://doi.org/10.3390/s22197274







