A Method for Medical Microscopic Images’ Sharpness Evaluation Based on NSST and Variance by Combining Time and Frequency Domains
Abstract
1. Introduction
- The NSST algorithm can decompose the image at multiple scales to obtain a low-frequency sub-band and several high-frequency sub-bands, and the image information contained in different sub-bands is also different. By calculating the variance of different sub-band coefficients, the interference caused by the background of the urine sediment image can be further reduced and the performance of the NSST algorithm can be improved, as to obtain a better clarity evaluation curve.
- Microscopic imaging technology will inevitably generate noise in images due to factors such as environment, equipment, and improper operation. In order to simulate the noise situation as much as possible, different noises are added to the experimental image, and a bilateral filter and a Gaussian filter are applied to the noise image to improve the noise resistance of the algorithm. Finally, the noise resistance of the improved NSST algorithm and other algorithms in this study are tested under the same operating conditions, with the results showing that the improved NSST algorithm has a better anti-noise performance than other algorithms used in this study.
2. Related Work
3. Materials and Methods
3.1. Non-Subsampled Shearlet Wave Transform (NSST)
3.2. Analyses of Algorithm Theories
3.3. Algorithm Improvement
3.4. Algorithm Implementation
- Perform NSST decomposition on an image to obtain one low-frequency sub-band and several high-frequency sub-bands.
- Obtain the variance processing coefficient of the low-frequency sub-band image in the NSST transform domain, as defined below:
- 3.
- Combine the energy of the high- and low-frequency components to calculate the sharpness evaluation value, as defined below:
| Algorithm 1. Pseudo-Code of the Algorithm |
| Input: N1 is the number of pictures to be processed. Output: H is the definition of the evaluation value.
|
3.5. Analysis of the Algorithm’s Performance
4. Results
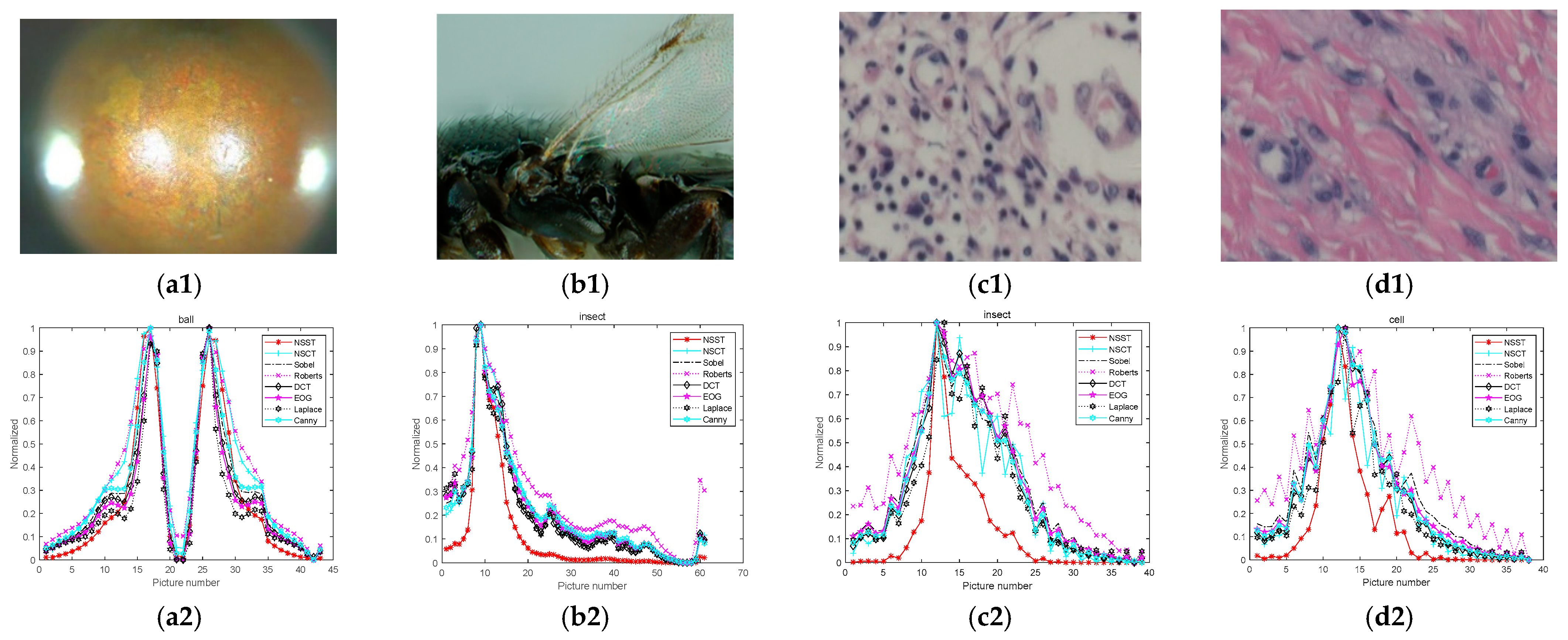
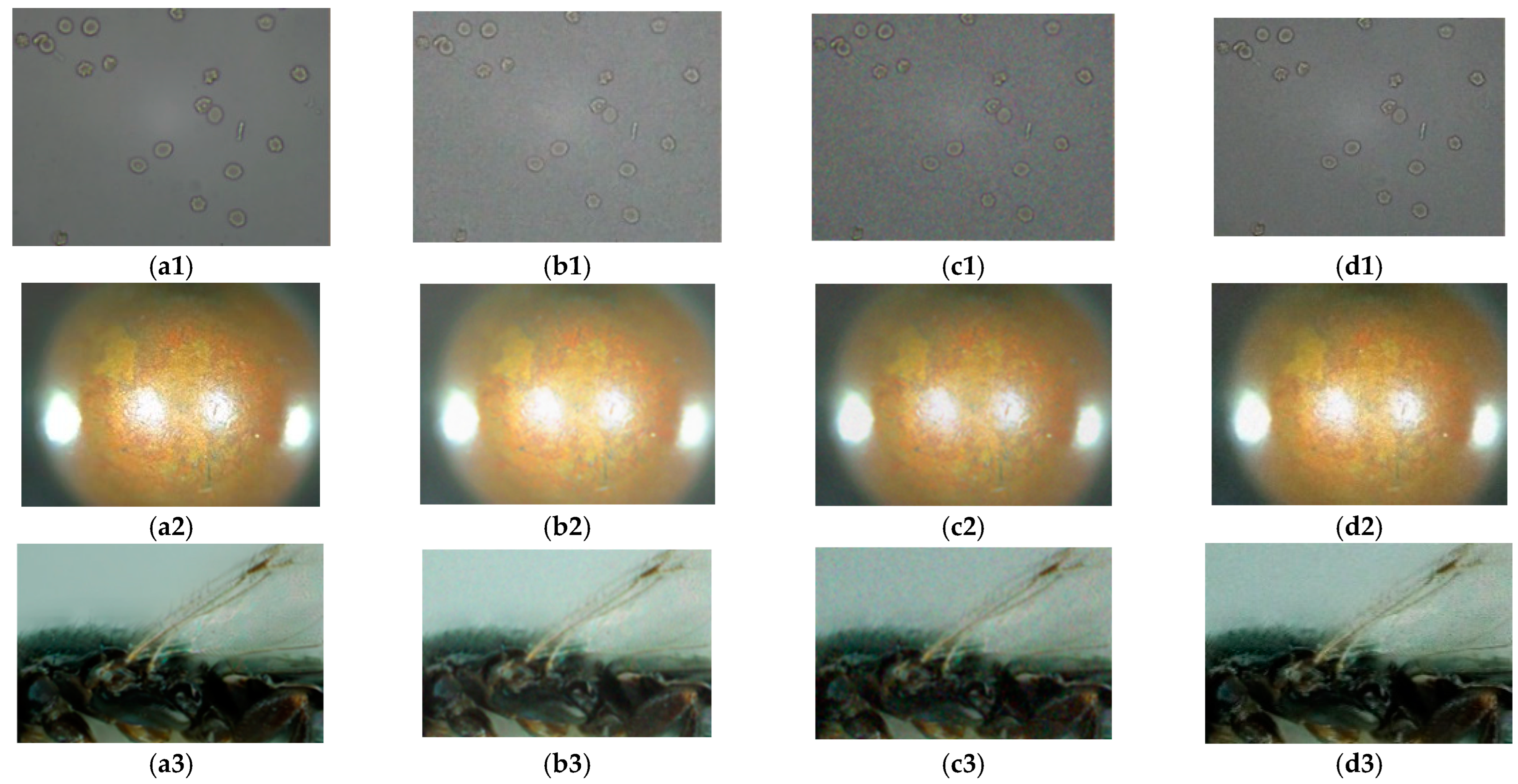
4.1. Analyses and Comparison of Algorithms
4.2. Noise Immunity Test
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nilanjan Dey, A.S.A.A. Digital Analysis of Microscopic Images in Medicine. J. Adv. Microsc. Res. 2015, 10, 1–13. [Google Scholar]
- Dogan, H.; Baykal, E.; Ekinci, M. Image panorama without loss of focusing for microscopic systems. In Proceedings of the 2017 Interna-tional Conference on Computer Science and Engineering (UBMK), Antalya, Turkey, 5–8 October 2017; IEEE, 2017. [Google Scholar]
- Qu, Y.; Zhu, S.; Zhang, P. A self-adaptive and nonmechanical motion autofocusing system for optical microscopes. Microsc. Res. Tech. 2016, 79, 1112–1122. [Google Scholar] [CrossRef]
- Xia, X.; Yao, Y.; Liang, J.; Fang, S.; Yang, Z.; Cui, D. Evaluation of focus measures for the autofocus of line scan cameras. Optik 2016, 127, 7762–7775. [Google Scholar] [CrossRef]
- Xia, X.; Yin, L.; Yao, Y.; Wu, W.; Zhang, Y. Combining two focus measures to improve performance. Meas. Sci. Technol. 2017, 28, 105401. [Google Scholar] [CrossRef]
- Liu, S.; Liu, M.; Yang, Z. An image auto-focusing algorithm for industrial image measurement. EURASIP J. Adv. Signal Process. 2016, 2016, 1. [Google Scholar] [CrossRef]
- Cabazos-Marín, A.R.; Álvarez-Borrego, J. Automatic focus and fusion image algorithm using nonlinear correlation: Image quality evaluation. Optik 2018, 164, 224–242. [Google Scholar] [CrossRef]
- Hore, S.; Chakraborty, S.; Ashour, A.; Dey, N.; Ashour, A.S.; Sifaki-Pistolla, D.; Bhattacharya, T.; Chaudhuri, S.R.B. Finding Contours of Hippocampus Brain Cell Using Microscopic Image Analysis. J. Adv. Microsc. Res. 2015, 10, 93–103. [Google Scholar] [CrossRef]
- Akiyama, A.; Kobayashi, N.; Mutoh, E.; Kumagai, H.; Yamada, H.; Ishii, H. Infrared image guidance for ground vehicle based on fast wavelet image focusing and tracking. In Proceedings of the Novel Optical Systems Design and Optimization XII, San Diego, CA, USA, 2–6 August 2009; pp. 50–57. [Google Scholar] [CrossRef]
- Makkapati, V.V. Improved wavelet-based microscope autofocusing for blood smears by using segmentation. In Proceedings of the 2009 IEEE International Conference on Automation Science and Engineering, Bangalore, India, 22–15 August 2009; IEEE, 2009; pp. 208–211. [Google Scholar] [CrossRef]
- Chen, R.; van Beek, P. Improving the accuracy and low-light performance of contrast-based autofocus using supervised machine learning. Pattern Recognit. Lett. 2015, 56, 30–37. [Google Scholar] [CrossRef]
- Matsui, S.; Nagahara, H.; Taniguchi, R.-I. Half-sweep imaging for depth from defocus. Image Vis. Comput. 2014, 32, 954–964. [Google Scholar] [CrossRef]
- Lv, M.; Wang, Z. Study on automatic focusing algorithm of optical microscope. China Meas. Test 2018, 44, 11–16. [Google Scholar] [CrossRef]
- Beagum, S.; Dey, N.; Ashour, A.S.; Sifaki-Pistolla, D.; Balas, V.E. Nonparametric de-noising filter optimization using structure-based microscopic image classification. Microsc. Res. Tech. 2016, 80, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Ashour, A.S.; Beagum, S.; Dey, N.; Ashour, A.S.; Pistolla, D.S.; Nguyen, G.N.; Le, D.-N.; Shi, F. Light microscopy image de-noising using optimized LPA-ICI filter. Neural Comput. Appl. 2018, 29, 1517–1533. [Google Scholar] [CrossRef]
- Shahdoosti, H.R.; Khayat, O. Image denoising using sparse representation classification and non-subsampled shearlet transform. Signal Image Video Process. 2016, 10, 1081–1087. [Google Scholar] [CrossRef]
- Zhan, K.; Shi, J.; Wang, H.; Xie, Y.; Li, Q. Computational Mechanisms of Pulse-Coupled Neural Networks: A Comprehensive Review. Arch. Comput. Methods Eng. 2016, 24, 573–588. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, H.; Ma, Y. A new auto-focus measure based on medium frequency discrete cosine transform filtering and discrete cosine transform. Appl. Comput. Harmon. Anal. 2016, 40, 430–437. [Google Scholar] [CrossRef]
- Liu, X.; Mei, W.; Du, H.; Bei, J. A novel image fusion algorithm based on nonsubsampled shearlet transform and morphological component analysis. Signal Image Video Process. 2015, 10, 959–966. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, S.; Wang, C.; Xu, J. Single image super-resolution via hybrid resolution NSST prediction. Comput. Vis. Image Underst. 2021, 207, 103202. [Google Scholar] [CrossRef]
- Diwakar, M.; Singh, P.; Shankar, A. Multi-modal medical image fusion framework using co-occurrence filter and local extrema in NSST domain. Biomed. Signal Process. Control 2021, 68, 102788. [Google Scholar] [CrossRef]
- Li, Y.; Po, L.M.; Xu, X.; Feng, L.; Yuan, F.; Cheung, C.-H.; Cheung, K.-W. No-reference image quality assessment with shearlet transform and deep neural networks. Neurocomputing 2015, 154, 94–109. [Google Scholar] [CrossRef]
- Peng, H.; Li, B.; Yang, Q.; Wang, J. Multi-focus image fusion approach based on CNP systems in NSCT domain. Comput. Vis. Image Underst. 2021, 210, 103228. [Google Scholar] [CrossRef]
- Liu, J.; Jia, Z.; Qin, X.; Yang, J.; Hu, Y. NSCT Remote Sensing Image Denoising Based on Threshold of Free Distributed FDR. Procedia Eng. 2011, 24, 616–620. [Google Scholar] [CrossRef]
- Da Cunha, A.; Zhou, J.; Do, M. The Nonsubsampled Contourlet Transform: Theory, Design, and Applications. IEEE Trans. Image Process. 2006, 15, 3089–3101. [Google Scholar] [CrossRef] [PubMed]
- Guorong, G.; Luping, X.; Dongzhu, F. Multi-focus image fusion based on non-subsampled shearlet transform. IET Image Processing 2013, 7, 633–639. [Google Scholar] [CrossRef]
- Gao, Y.; Ma, S.; Liu, J.; Liu, Y.; Zhang, X. Fusion of medical images based on salient features extraction by PSO optimized fuzzy logic in NSST do-main. Biomed. Signal Processing Control. 2021, 69, 102852. [Google Scholar] [CrossRef]
- He, C.; Li, X.; Hu, Y.; Ye, Z.; Kang, H. Microscope images automatic focus algorithm based on eight-neighborhood operator and least square planar fitting. Optik 2020, 206, 164232. [Google Scholar] [CrossRef]
- Lin, Z.; Huang, C.; Lu, A. Autofocus method based on blur difference qualitative analysis. J. Comput. Appli-Cations 2015, 35, 2969–2973+2979. [Google Scholar]
- Lüthi, B.; Thomas, N.; Hviid, S.; Rueffer, P. An efficient autofocus algorithm for a visible microscope on a Mars lander. Planet. Space Sci. 2010, 58, 1258–1264. [Google Scholar] [CrossRef]
- Chu, F.; Mao, Y.; Zeng, J.; Bian, Z.; Hu, A.; Wen, H. Application of auto-focus algorithm in welding pool imaging system. Weld. World 2022, 25, 1–12. [Google Scholar] [CrossRef]
- Yao, Y.; Abidi, B.; Doggaz, N.; Abidi, M. Evaluation of sharpness measures and search algorithms for the auto focusing of high-magnification images. In Proceedings of the Visual Information Processing XV, Orlando, FL, USA, 17 April 2006; SPIE, 2006. [Google Scholar]
- Zhang, Z.; Liu, Y.; Xiong, Z.; Li, J.; Zhang, M. Focus and Blurriness Measure Using Reorganized DCT Coefficients for an Autofocus Application. IEEE Trans. Circuits Syst. Video Technol. 2016, 28, 15–30. [Google Scholar] [CrossRef]
- Bilen, H.; Hocaoglu, M.A.; Unel, M.; Sabanovic, A. Developing robust vision modules for microsystems applications. Mach. Vis. Appl. 2010, 23, 25–42. [Google Scholar] [CrossRef]
- Huang, M.; Liu, Y.; Yang, Y. Edge detection of ore and rock on the surface of explosion pile based on improved Canny operator. Alex. Eng. J. 2022, 61, 10769–10777. [Google Scholar] [CrossRef]
- Chen, D.; Cheng, J.-J.; He, H.-Y.; Ma, C.; Yao, L.; Jin, C.-B.; Cao, Y.-S.; Li, J.; Ji, P. Computed tomography reconstruction based on canny edge detection algorithm for acute expansion of epidural hematoma. J. Radiat. Res. Appl. Sci. 2022, 15, 279–284. [Google Scholar] [CrossRef]
- Helmli, F.; Scherer, S. Adaptive shape from focus with an error estimation in light microscopy. In Proceedings of the 2nd International Symposium on Image and Signal Processing and Analysis, Pula, Croatia, 19–21 June 2001; pp. 2319–212001. [Google Scholar] [CrossRef]
- Hurtado-Pérez, R.; Toxqui-Quitl, C.; Padilla-Vivanco, A.; Aguilar-Valdez, J.F.; Ortega-Mendoza, G. Focus measure method based on the modulus of the gradient of the color planes for digital microscopy. Opt. Eng. 2018, 57, 023106. [Google Scholar] [CrossRef]
- AS, R.A.; Gopalan, S. Comparative Analysis of Eight Direction Sobel Edge Detection Algorithm for Brain Tumor MRI Images. Procedia Comput. Sci. 2022, 201, 487–494. [Google Scholar]
- Joshi, R.; Zaman, A.; Katkoori, S. Fast Sobel Edge Detection for IoT Edge Devices. SN Comput. Sci. 2022, 3, 1–13. [Google Scholar] [CrossRef]
- Redondo, R.; Bueno, G.; Valdiviezo, J.C.; Nava, R.; Cristobal, G.; Deniz, O.; García-Rojo, M.; Salido, J.; Fernández, M.D.M.; Vidal, J.; et al. Autofocus evaluation for brightfield microscopy pathology. J. Biomed. Opt. 2012, 17, 0360088. [Google Scholar] [CrossRef]










| Algorithms | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | Time |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NSCT | 0.4545 | 0.3771 | 0.3238 | 0.2672 | 0.4120 | 0.3974 | 0.4322 | 0.3541 | 0.2642 | 0.4577 | 0.3778 | 135.54 |
| Sobel | 0.2433 | 0.2699 | 0.2398 | 0.2853 | 0.3552 | 0.2696 | 0.3659 | 0.1550 | 0.3197 | 0.2915 | 0.2473 | 2.40 |
| Roberts | 0.2247 | 0.2254 | 0.2249 | 0.2861 | 0.2879 | 0.2823 | 0.2019 | 0.2009 | 0.2994 | 0.2366 | 0.2423 | 2.33 |
| DCT | 0.2631 | 0.2872 | 0.2906 | 0.2518 | 0.4054 | 0.2662 | 0.3717 | 0.1876 | 0.4235 | 0.2989 | 0.2759 | 10.98 |
| EOG | 0.2635 | 0.3048 | 0.2972 | 0.3224 | 0.3537 | 0.2431 | 0.1902 | 0.1790 | 0.3892 | 0.2819 | 0.2929 | 2.34 |
| Laplacian | 0.2874 | 0.3093 | 0.2959 | 0.3639 | 0.2859 | 0.2139 | 0.1614 | 0.2420 | 0.4768 | 0.2274 | 0.2666 | 2.34 |
| Canny | 0.2733 | 0.2981 | 0.3116 | 0.2766 | 0.3921 | 0.2625 | 0.2123 | 0.1875 | 0.2985 | 0.3092 | 0.2703 | 9.36 |
| NSST | 0.4258 | 0.5170 | 0.4500 | 0.3248 | 0.4899 | 0.4224 | 0.4417 | 0.3587 | 0.3478 | 0.5029 | 0.5263 | 33.97 |
| Algorithms | Ball (Left Peak) | Ball (Right Peak) | Insect | Cells | Time |
|---|---|---|---|---|---|
| NSCT | 0.4318 | 0.4026 | 0.4085 | 0.1897 | 394.67 |
| Sobel | 0.4361 | 0.3454 | 0.2667 | 0.1304 | 5.19 |
| Roberts | 0.3586 | 0.2988 | 0.3245 | 0.2042 | 5.95 |
| DCT | 0.2412 | 0.2111 | 0.3131 | 0.1970 | 31.79 |
| EOG | 0.4473 | 0.3889 | 0.2441 | 0.2288 | 5.96 |
| Laplacian | 0.4492 | 0.4078 | 0.2586 | 0.2429 | 5.97 |
| Canny | 0.4393 | 0.3780 | 0.2738 | 0.2653 | 52.98 |
| NSST | 0.4719 | 0.4196 | 0.3956 | 0.3146 | 77.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Zhou, H.; Yu, H.; Hu, R.; Zhang, G.; Hu, J.; He, T. A Method for Medical Microscopic Images’ Sharpness Evaluation Based on NSST and Variance by Combining Time and Frequency Domains. Sensors 2022, 22, 7607. https://doi.org/10.3390/s22197607
Wu X, Zhou H, Yu H, Hu R, Zhang G, Hu J, He T. A Method for Medical Microscopic Images’ Sharpness Evaluation Based on NSST and Variance by Combining Time and Frequency Domains. Sensors. 2022; 22(19):7607. https://doi.org/10.3390/s22197607
Chicago/Turabian StyleWu, Xuecheng, Houkui Zhou, Huimin Yu, Roland Hu, Guangqun Zhang, Junguo Hu, and Tao He. 2022. "A Method for Medical Microscopic Images’ Sharpness Evaluation Based on NSST and Variance by Combining Time and Frequency Domains" Sensors 22, no. 19: 7607. https://doi.org/10.3390/s22197607
APA StyleWu, X., Zhou, H., Yu, H., Hu, R., Zhang, G., Hu, J., & He, T. (2022). A Method for Medical Microscopic Images’ Sharpness Evaluation Based on NSST and Variance by Combining Time and Frequency Domains. Sensors, 22(19), 7607. https://doi.org/10.3390/s22197607






