Detection of Diabetic Retinopathy Using Extracted 3D Features from OCT Images
Abstract
:1. Introduction
1.1. Related Work
1.2. Limitation of the Existing Works and Proposed Method
- Most of the techniques do not consider the 3D characteristics of the images and are limited to the 2D image features.
- A significant quantity of data is needed to train and test deep learning models, which may not be available.
- The paper is the first of its kind to analyze the 3D retinal layer by using low-level (first-order reflectivity) and high-level (3D thickness) information.
- Backpropagated neural networks are optimized to combine low-level and high-level information to further improve performance.
- Comparing the suggested approach to related methods, it performs better.
2. Materials and Methods
2.1. Patient Data
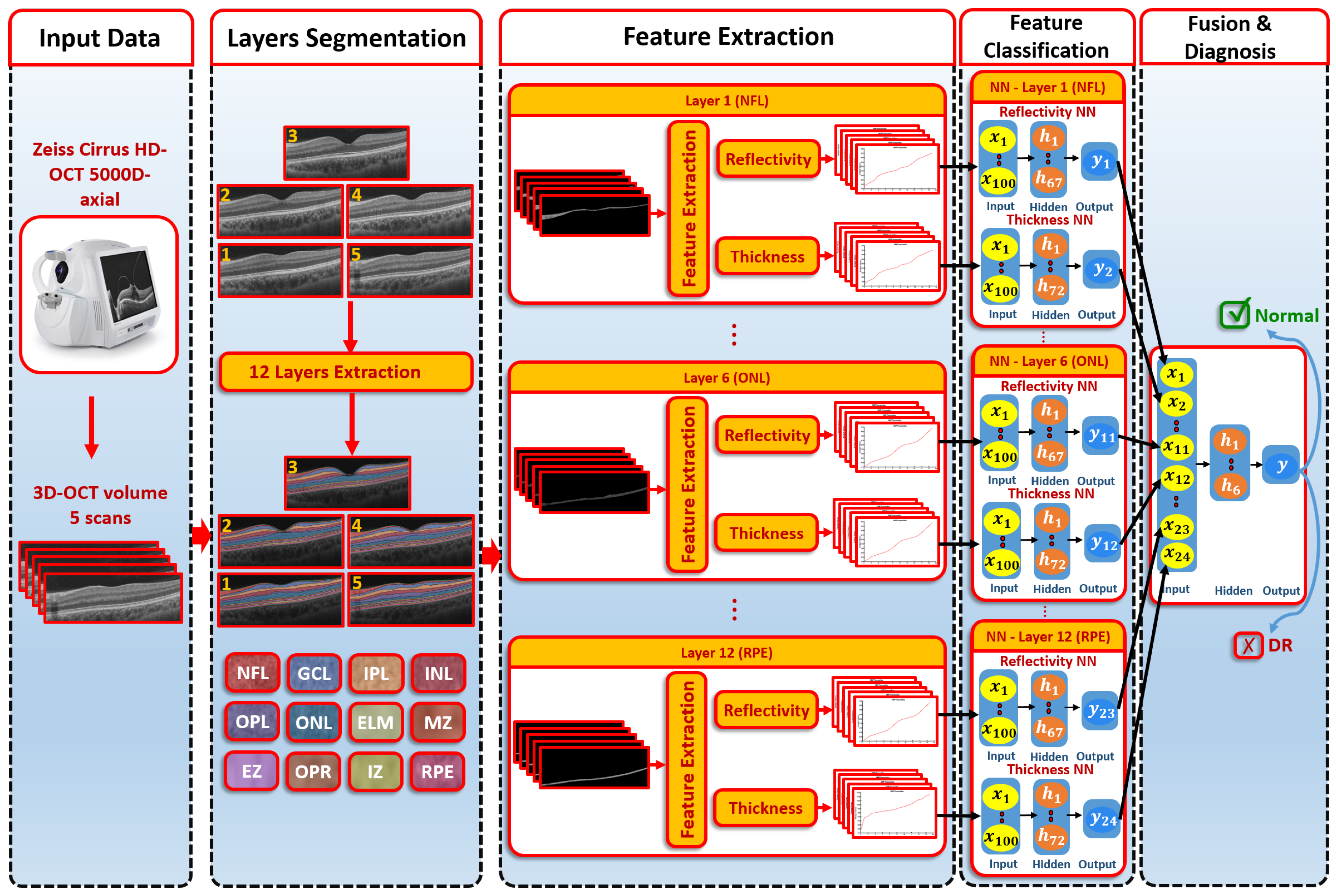
2.2. Proposed Computer-Aided Diagnostic (CAD) System
2.2.1. Segmentation of 3D-OCT Images
- The B-scan OCT was aligned to a shape data formed by an expert. These shape data contained manual segmentations of the area in the center of the macula (foveal) of normal and diseased retina shape priors.
- The central B-scan was divided into twelve distinct layers based on intensity, MGRF spatial interactions, and shape.
- A nonrigid deformation B-scan was used as a prior shape pattern in each segmented B-scan process.
- The models of shape prior were repeated in each slice to get the final 3D segmentation.
2.2.2. Feature Extraction
- Locate the surface of the target and reference objects.
- Initially: adjust the minimum and maximum potential at the corresponding reference surface and the target surface, respectively.
- Using Equation (3), between both isosurfaces, can be estimated.
- Repeat the third step until convergence is achieved (i.e., no change occurs in the evaluated values between the iterations).
2.2.3. Classification System
- The first-order reflectivity feature of each layer was used to feed neural networks (NNs), each was composed of one hidden layer that involved 67 neurons. Each NN was applied to each layer individually. Through each NN, each layer gave a probability between 0 and 1
- The 3D thickness feature was fed to the NNs. Each NN was composed of one hidden layer containing 72 neurons and applied to each layer individually. The output of each NN was the probability of each layer.
- In the second stage, we fused the probabilities resulting from the previous first-stage NNs. The second-stage NN contained one hidden layer of 6 neurons. The output represented the final diagnosis.
- At first, the weights of NNs were initialized randomly.
- All outputs in hidden layers and output layers for neurons were calculated.
- The activation function was applied on each neuron of the outputs calculated in step 2.
- By using the backpropagation approach, the different weights were updated.
- Steps 2, 3, and 4 were replicated until the weights became stable.
2.2.4. System Evaluation
3. Results
3.1. Results in the Case of Using the Reflectivity Only
3.2. Results in the Case of Using the Thickness Only
3.3. Results of the Proposed System
3.4. Comparison Results to Other ML Methods
3.5. Comparison Results to DL Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Radomska-Leśniewska, D.M.; Osiecka-Iwan, A.; Hyc, A.; Góźdź, A.; Dąbrowska, A.M.; Skopiński, P. Therapeutic potential of curcumin in eye diseases. Cent. Eur. J. Immunol. 2019, 44, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.; Wong, T.Y. Obesity and eye diseases. Surv. Ophthalmol. 2007, 52, 180–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, T.; Jayasudha, R.; Chakravarthy, S.; Prashanthi, G.S.; Bhargava, A.; Tyagi, M.; Rani, P.K.; Pappuru, R.R.; Sharma, S.; Shivaji, S. Alterations in the gut bacterial microbiome in people with type 2 diabetes mellitus and diabetic retinopathy. Sci. Rep. 2021, 11, 2738. [Google Scholar] [CrossRef]
- Peplow, P.; Martinez, B. MicroRNAs as biomarkers of diabetic retinopathy and disease progression. Neural Regen. Res. 2019, 14, 1858. [Google Scholar] [CrossRef]
- Oh, K.; Kang, H.M.; Leem, D.; Lee, H.; Seo, K.Y.; Yoon, S. Early detection of diabetic retinopathy based on deep learning and ultra-wide-field fundus images. Sci. Rep. 2021, 11, 1897. [Google Scholar] [CrossRef] [PubMed]
- Teo, Z.L.; Tham, Y.C.; Yu, M.; Chee, M.L.; Rim, T.H.; Cheung, N.; Bikbov, M.M.; Wang, Y.X.; Tang, Y.; Lu, Y.; et al. Global Prevalence of Diabetic Retinopathy and Projection of Burden through 2045. Ophthalmology 2021, 128, 1580–1591. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.; Wong, T.Y. Management Paradigms for Diabetic Macular Edema. Am. J. Ophthalmol. 2014, 157, 505–513.e8. [Google Scholar] [CrossRef]
- Antoszyk, A.N.; Glassman, A.R.; Beaulieu, W.T.; Jampol, L.M.; Jhaveri, C.D.; Punjabi, O.S.; Salehi-Had, H.; Wells, J.A.; Maguire, M.G.; Stockdale, C.R.; et al. Effect of Intravitreous Aflibercept vs. Vitrectomy With Panretinal Photocoagulation on Visual Acuity in Patients With Vitreous Hemorrhage From Proliferative Diabetic Retinopathy. JAMA 2020, 324, 2383. [Google Scholar] [CrossRef]
- Everett, L.A.; Paulus, Y.M. Laser Therapy in the Treatment of Diabetic Retinopathy and Diabetic Macular Edema. Curr. Diabetes Rep. 2021, 21, 35. [Google Scholar] [CrossRef]
- Maniadakis, N.; Konstantakopoulou, E. Cost Effectiveness of Treatments for Diabetic Retinopathy: A Systematic Literature Review. Pharmacoeconomics 2019, 37, 995–1010. [Google Scholar] [CrossRef]
- Witkin, A.; Salz, D. Imaging in diabetic retinopathy. Middle East Afr. J. Ophthalmol. 2015, 22, 145. [Google Scholar] [CrossRef] [PubMed]
- Lains, I.; Wang, J.C.; Cui, Y.; Katz, R.; Vingopoulos, F.; Staurenghi, G.; Vavvas, D.G.; Miller, J.W.; Miller, J.B. Retinal applications of swept source optical coherence tomography (OCT) and optical coherence tomography angiography (OCTA). Prog. Retin. Eye Res. 2021, 84, 100951. [Google Scholar] [CrossRef] [PubMed]
- Priya, R.; Aruna, P. Diagnosis of diabetic retinopathy using machine learning techniques. ICTACT J. Soft Comput. 2013, 3, 563–575. [Google Scholar] [CrossRef]
- Foeady, A.Z.; Novitasari, D.C.R.; Asyhar, A.H.; Firmansjah, M. Automated Diagnosis System of Diabetic Retinopathy Using GLCM Method and SVM Classifier. In Proceedings of the 2018 5th International Conference on Electrical Engineering, Computer Science and Informatics (EECSI), Malang, Indonesia, 16–18 October 2018. [Google Scholar] [CrossRef]
- Abdelmaksoud, E.; El-Sappagh, S.; Barakat, S.; Abuhmed, T.; Elmogy, M. Automatic Diabetic Retinopathy Grading System Based on Detecting Multiple Retinal Lesions. IEEE Access 2021, 9, 15939–15960. [Google Scholar] [CrossRef]
- Rajput, G.; Reshmi, B.; Rajesh, I. Automatic detection and grading of diabetic maculopathy using fundus images. Procedia Comput. Sci. 2020, 167, 57–66. [Google Scholar] [CrossRef]
- Zhou, W.; Wu, C.; Yu, X. Computer aided diagnosis for diabetic retinopathy based on fundus image. In Proceedings of the 2018 37th Chinese Control Conference (CCC), Wuhan, China, 25–27 July 2018; pp. 9214–9219. [Google Scholar]
- Rahim, S.S.; Palade, V.; Shuttleworth, J.; Jayne, C. Automatic screening and classification of diabetic retinopathy and maculopathy using fuzzy image processing. Brain Inform. 2016, 3, 249–267. [Google Scholar] [CrossRef]
- Sánchez, C.I.; Niemeijer, M.; Dumitrescu, A.V.; Suttorp-Schulten, M.S.; Abramoff, M.D.; van Ginneken, B. Evaluation of a computer-aided diagnosis system for diabetic retinopathy screening on public data. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4866–4871. [Google Scholar] [CrossRef] [Green Version]
- Mansour, R.F. Deep-learning-based automatic computer-aided diagnosis system for diabetic retinopathy. Biomed. Eng. Lett. 2018, 8, 41–57. [Google Scholar] [CrossRef]
- Li, F.; Liu, Z.; Chen, H.; Jiang, M.; Zhang, X.; Wu, Z. Automatic detection of diabetic retinopathy in retinal fundus photographs based on deep learning algorithm. Transl. Vis. Sci. Technol. 2019, 8, 4. [Google Scholar] [CrossRef]
- Abbas, Q.; Fondon, I.; Sarmiento, A.; Jiménez, S.; Alemany, P. Automatic recognition of severity level for diagnosis of diabetic retinopathy using deep visual features. Med Biol. Eng. Comput. 2017, 55, 1959–1974. [Google Scholar] [CrossRef]
- Khalifa, N.E.M.; Loey, M.; Taha, M.H.N.; Mohamed, H.N.E.T. Deep transfer learning models for medical diabetic retinopathy detection. Acta Inform. Medica 2019, 27, 327. [Google Scholar] [CrossRef] [PubMed]
- Kassani, S.H.; Kassani, P.H.; Khazaeinezhad, R.; Wesolowski, M.J.; Schneider, K.A.; Deters, R. Diabetic retinopathy classification using a modified xception architecture. In Proceedings of the 2019 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT), Ajman, United Arab Emirates, 10–12 December 2019; pp. 1–6. [Google Scholar]
- Rahhal, D.; Alhamouri, R.; Albataineh, I.; Duwairi, R. Detection and Classification of Diabetic Retinopathy Using Artificial Intelligence Algorithms. In Proceedings of the 2022 13th International Conference on Information and Communication Systems (ICICS), Irbid, Jordan, 21–23 June 2022; pp. 15–21. [Google Scholar]
- Ibrahim, M.R.; Fathalla, K.M.; M. Youssef, S. HyCAD-OCT: A hybrid computer-aided diagnosis of retinopathy by optical coherence tomography integrating machine learning and feature maps localization. Appl. Sci. 2020, 10, 4716. [Google Scholar] [CrossRef]
- Ghazal, M.; Ali, S.S.; Mahmoud, A.H.; Shalaby, A.M.; El-Baz, A. Accurate Detection of Non-Proliferative Diabetic Retinopathy in Optical Coherence Tomography Images Using Convolutional Neural Networks. IEEE Access 2020, 8, 34387–34397. [Google Scholar] [CrossRef]
- ElTanboly, A.; Ismail, M.; Shalaby, A.; Switala, A.; El-Baz, A.; Schaal, S.; Gimel’farb, G.; El-Azab, M. A computer-aided diagnostic system for detecting diabetic retinopathy in optical coherence tomography images. Med. Phys. 2017, 44, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, H.S.; Eltanboly, A.; Shalaby, A.; Keynton, R.S.; Schaal, S.; El-Baz, A. Automated Diagnosis and Grading of Diabetic Retinopathy Using Optical Coherence Tomography. Investig. Opthalmol. Vis. Sci. 2018, 59, 3155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Shen, L.; Shen, M.; Tan, F.; Qiu, C.S. Deep learning based early stage diabetic retinopathy detection using optical coherence tomography. Neurocomputing 2019, 369, 134–144. [Google Scholar] [CrossRef]
- Aldahami, M.; Alqasemi, U. Classification of OCT Images for Detecting Diabetic Retinopathy Disease Using Machine Learning. 2020. Available online: https://assets.researchsquare.com/files/rs-47495/v1/bda4d133-29c5-4c2e-ae8d-f0d9cd084989.pdf?c=1642522911 (accessed on 19 September 2022).
- Sharafeldeen, A.; Elsharkawy, M.; Khalifa, F.; Soliman, A.; Ghazal, M.; AlHalabi, M.; Yaghi, M.; Alrahmawy, M.; Elmougy, S.; Sandhu, H.S.; et al. Precise higher-order reflectivity and morphology models for early diagnosis of diabetic retinopathy using OCT images. Sci. Rep. 2021, 11, 4730. [Google Scholar] [CrossRef]
- Eladawi, N.; Elmogy, M.; Khalifa, F.; Ghazal, M.; Ghazi, N.; Aboelfetouh, A.; Riad, A.; Sandhu, H.; Schaal, S.; El-Baz, A. Early diabetic retinopathy diagnosis based on local retinal blood vessel analysis in optical coherence tomography angiography (OCTA) images. Med. Phys. 2018, 45, 4582–4599. [Google Scholar] [CrossRef]
- Heisler, M.; Karst, S.; Lo, J.; Mammo, Z.; Yu, T.; Warner, S.; Maberley, D.; Beg, M.F.; Navajas, E.V.; Sarunic, M.V. Ensemble deep learning for diabetic retinopathy detection using optical coherence tomography angiography. Transl. Vis. Sci. Technol. 2020, 9, 20. [Google Scholar] [CrossRef]
- Le, D.; Alam, M.; Yao, C.K.; Lim, J.I.; Hsieh, Y.T.; Chan, R.V.; Toslak, D.; Yao, X. Transfer learning for automated OCTA detection of diabetic retinopathy. Transl. Vis. Sci. Technol. 2020, 9, 35. [Google Scholar] [CrossRef]
- Sandhu, H.S.; Elmogy, M.; Sharafeldeen, A.T.; Elsharkawy, M.; El-Adawy, N.; Eltanboly, A.; Shalaby, A.; Keynton, R.; El-Baz, A. Automated Diagnosis of Diabetic Retinopathy Using Clinical Biomarkers, Optical Coherence Tomography, and Optical Coherence Tomography Angiography. Am. J. Ophthalmol. 2020, 216, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Sleman, A.A.; Soliman, A.; Elsharkawy, M.; Giridharan, G.; Ghazal, M.; Sandhu, H.; Schaal, S.; Keynton, R.; Elmaghraby, A.; El-Baz, A. A novel 3D segmentation approach for extracting retinal layers from optical coherence tomography images. Med. Phys. 2021, 48, 1584–1595. [Google Scholar] [CrossRef] [PubMed]
- Soliman, A.; Khalifa, F.; Dunlap, N.; Wang, B.; Abou El-Ghar, M.; El-Baz, A. An iso-surfaces based local deformation handling framework of lung tissues. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1253–1259. [Google Scholar]
- Powers, D.M. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv 2020, arXiv:2010.16061. [Google Scholar]
- Sandhu, H.S.; Eladawi, N.; Elmogy, M.; Keynton, R.; Helmy, O.; Schaal, S.; El-Baz, A. Automated diabetic retinopathy detection using optical coherence tomography angiography: A pilot study. Br. J. Ophthalmol. 2018, 102, 1564–1569. [Google Scholar] [CrossRef]

| Model | Reflectivity | Thickness | Proposed System | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sens. | Spec. | Acc. | Sens. | Spec. | Acc. | Sens. | Spec. | Acc. | ||
| 5 Folds | Layer 1 | |||||||||
| Layer 2 | ||||||||||
| Layer 3 | ||||||||||
| Layer 4 | ||||||||||
| Layer 5 | ||||||||||
| Layer 6 | ||||||||||
| Layer 7 | ||||||||||
| Layer 8 | ||||||||||
| Layer 9 | ||||||||||
| Layer 10 | ||||||||||
| Layer 11 | ||||||||||
| Layer 12 | ||||||||||
| Fusion | ||||||||||
| 10 Folds | Layer 1 | |||||||||
| Layer 2 | ||||||||||
| Layer 3 | ||||||||||
| Layer 4 | ||||||||||
| Layer 5 | ||||||||||
| Layer 6 | ||||||||||
| Layer 7 | ||||||||||
| Layer 8 | ||||||||||
| Layer 9 | ||||||||||
| Layer 10 | ||||||||||
| Layer 11 | ||||||||||
| Layer 12 | ||||||||||
| Fusion | ||||||||||
| LOSO | Layer 1 | |||||||||
| Layer 2 | ||||||||||
| Layer 3 | ||||||||||
| Layer 4 | ||||||||||
| Layer 5 | ||||||||||
| Layer 6 | ||||||||||
| Layer 7 | ||||||||||
| Layer 8 | ||||||||||
| Layer 9 | ||||||||||
| Layer 10 | ||||||||||
| Layer 11 | ||||||||||
| Layer 12 | ||||||||||
| Fusion | ||||||||||
| Classifier | Sens. | Spec. | Acc. | |
|---|---|---|---|---|
| 5 Folds | SVM | |||
| KNN | ||||
| DT | ||||
| NB | ||||
| Proposed System | ||||
| 10 Folds | SVM | |||
| KNN | ||||
| DT | ||||
| NB | ||||
| Proposed System | ||||
| LOSO | SVM | |||
| KNN | ||||
| DT | ||||
| NB | ||||
| Proposed System | ||||
| Classifier | Sens. | Spec. | Acc. | |
|---|---|---|---|---|
| 5 Folds | Google Net | |||
| Resnet-50 | ||||
| Proposed System | ||||
| 10 Folds | Google Net | |||
| Resnet-50 | ||||
| Proposed System | ||||
| Related Work | Accuracy % |
|---|---|
| ElTanboly et al. [28], 2017 | 92.0 |
| Sandhu et al. [40], 2018 | 94.3 |
| Sandhu et al. [29], 2018 | 95.0 |
| Li et al. [30], 2019 | 92.0 |
| Ghazal et al. [27], 2020 | 94.0 |
| Proposed System | 96.8 |
| Data Type | Sens. | Spec. | Acc. | |
|---|---|---|---|---|
| 5 Folds | 2D | |||
| Proposed System (3D) | ||||
| 10 Folds | 2D | |||
| Proposed System (3D) | ||||
| LOSO | 2D | |||
| Proposed System (3D) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elgafi, M.; Sharafeldeen, A.; Elnakib, A.; Elgarayhi, A.; Alghamdi, N.S.; Sallah, M.; El-Baz, A. Detection of Diabetic Retinopathy Using Extracted 3D Features from OCT Images. Sensors 2022, 22, 7833. https://doi.org/10.3390/s22207833
Elgafi M, Sharafeldeen A, Elnakib A, Elgarayhi A, Alghamdi NS, Sallah M, El-Baz A. Detection of Diabetic Retinopathy Using Extracted 3D Features from OCT Images. Sensors. 2022; 22(20):7833. https://doi.org/10.3390/s22207833
Chicago/Turabian StyleElgafi, Mahmoud, Ahmed Sharafeldeen, Ahmed Elnakib, Ahmed Elgarayhi, Norah S. Alghamdi, Mohammed Sallah, and Ayman El-Baz. 2022. "Detection of Diabetic Retinopathy Using Extracted 3D Features from OCT Images" Sensors 22, no. 20: 7833. https://doi.org/10.3390/s22207833
APA StyleElgafi, M., Sharafeldeen, A., Elnakib, A., Elgarayhi, A., Alghamdi, N. S., Sallah, M., & El-Baz, A. (2022). Detection of Diabetic Retinopathy Using Extracted 3D Features from OCT Images. Sensors, 22(20), 7833. https://doi.org/10.3390/s22207833










