A Deep Residual Neural Network for Image Reconstruction in Biomedical 3D Magnetic Induction Tomography
Abstract
1. Introduction
2. Materials and Methods
2.1. The Forward Problem
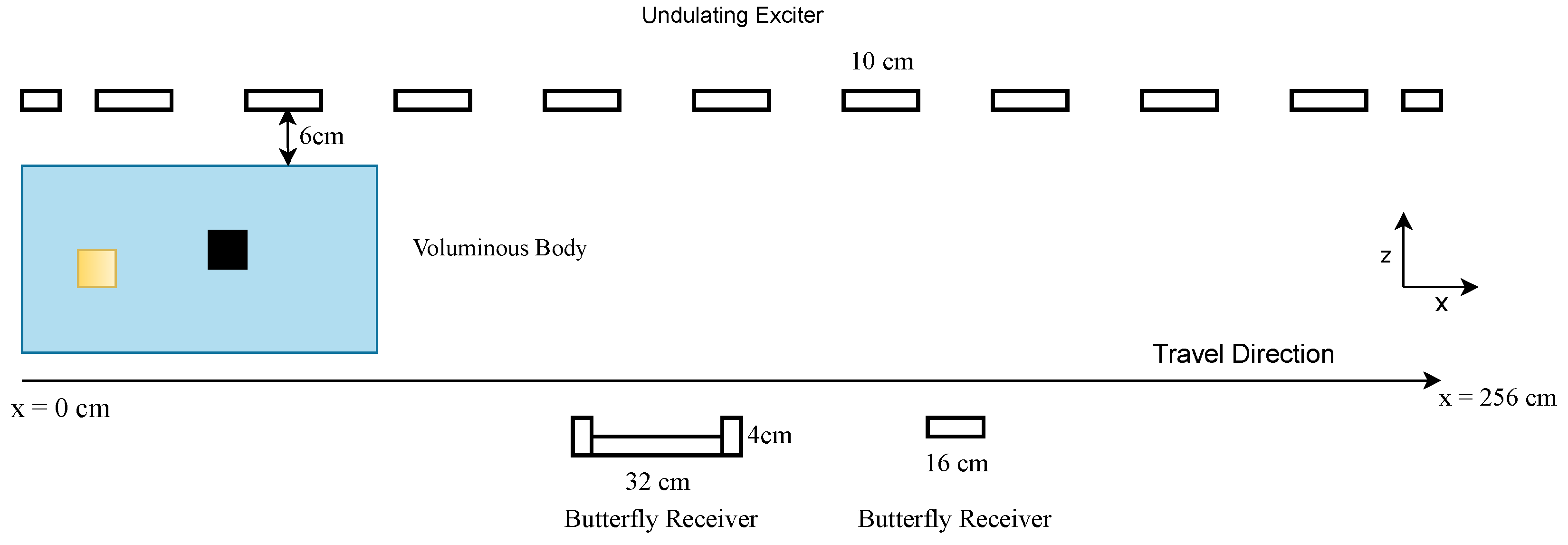
2.1.1. MIT Setup
2.1.2. Theory of the Forward Problem
2.2. The Inverse Problem
2.3. Dataset
Data Preprocessing
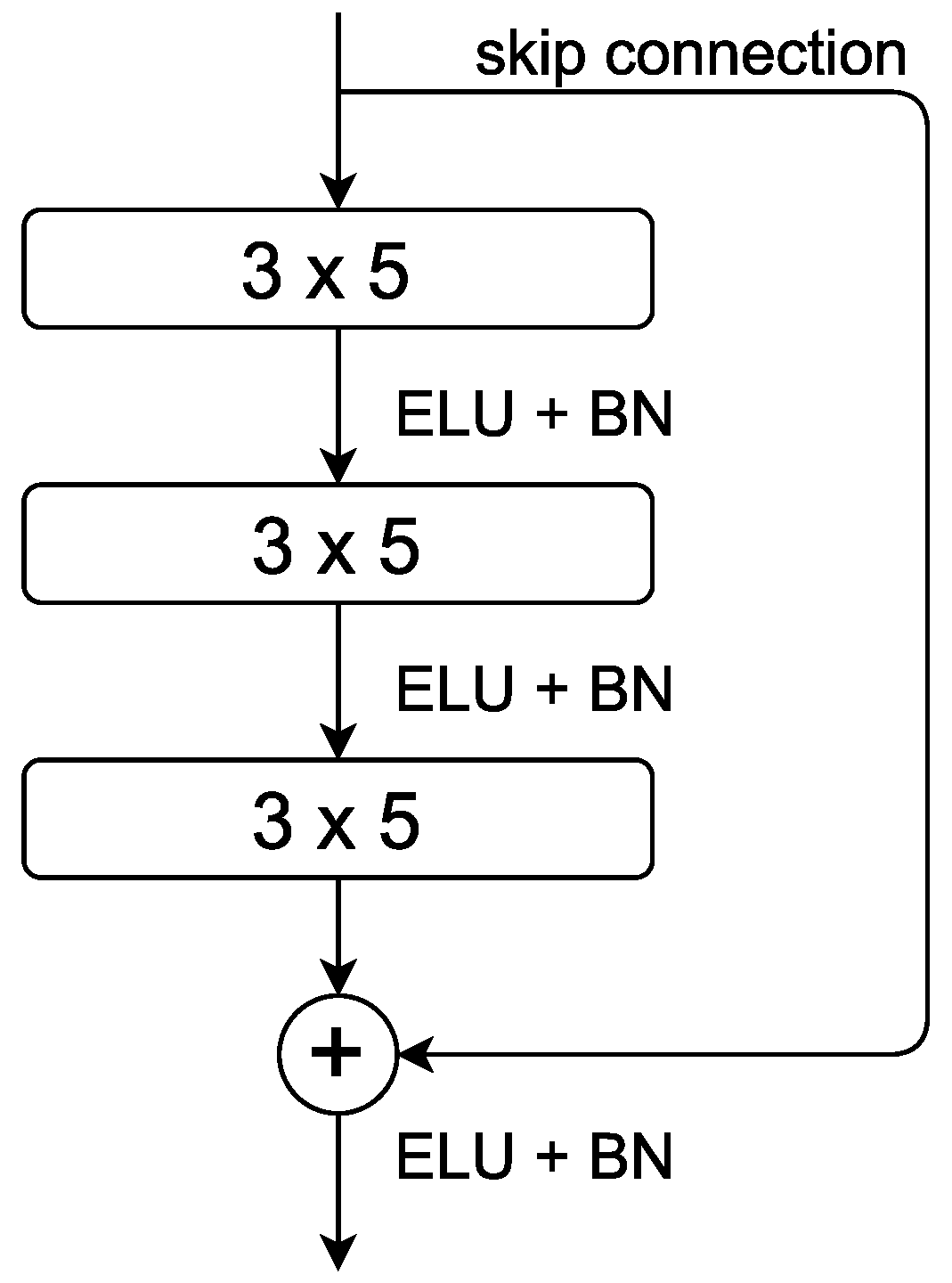
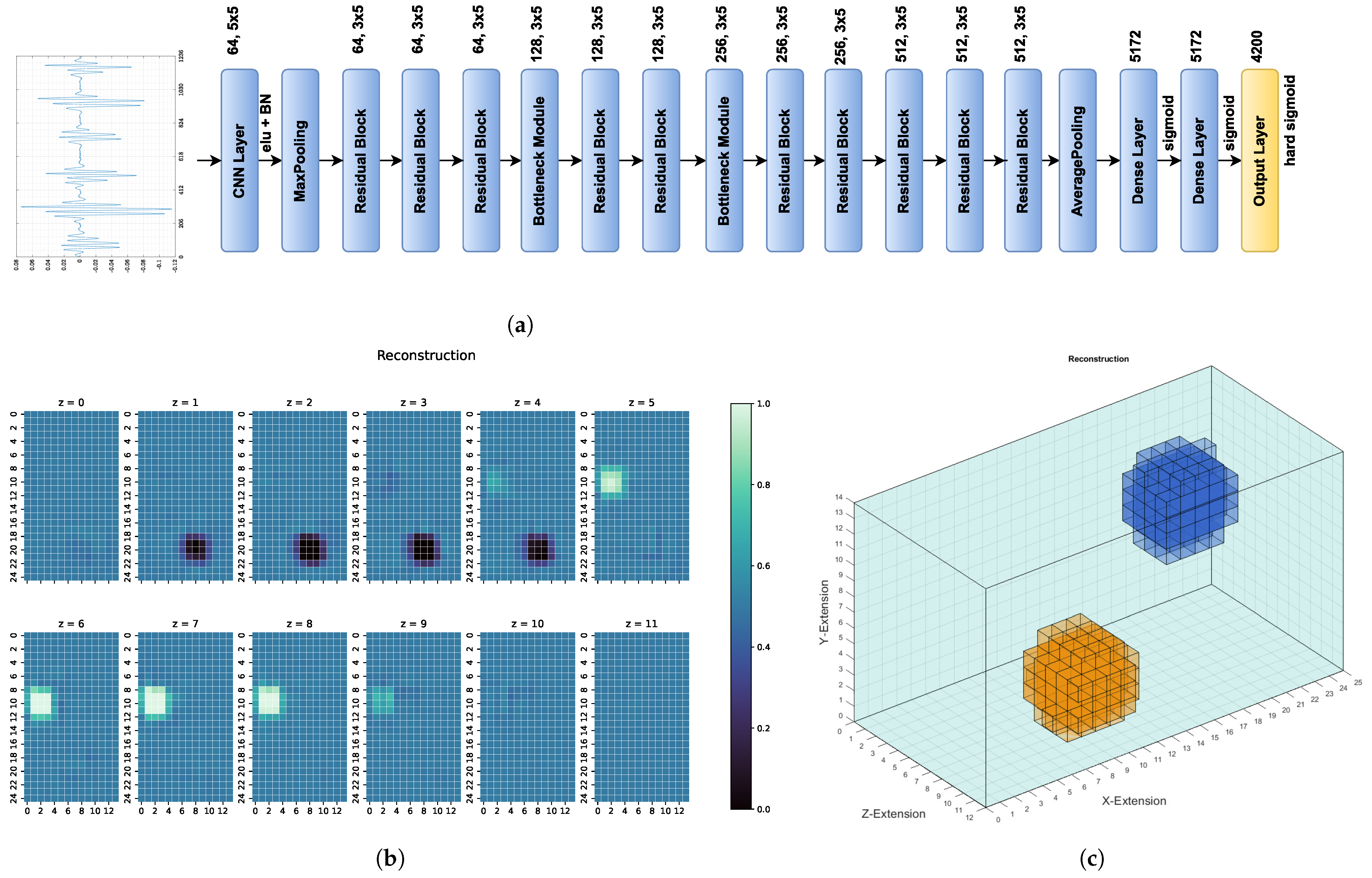
2.4. Structure of ResNet
2.5. Initialisation and Training of ResNet
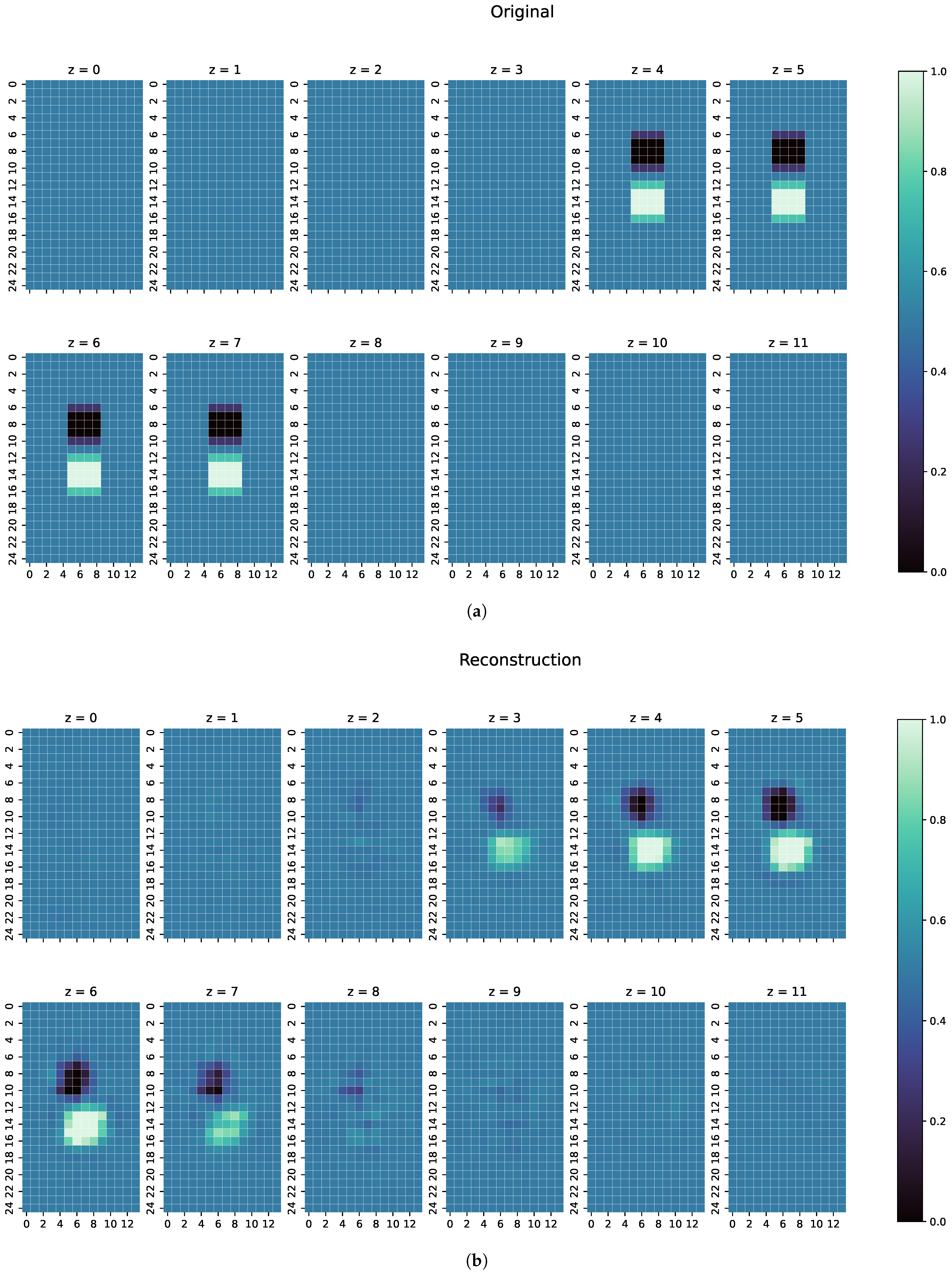
3. Results
3.1. Metrics
3.2. Results
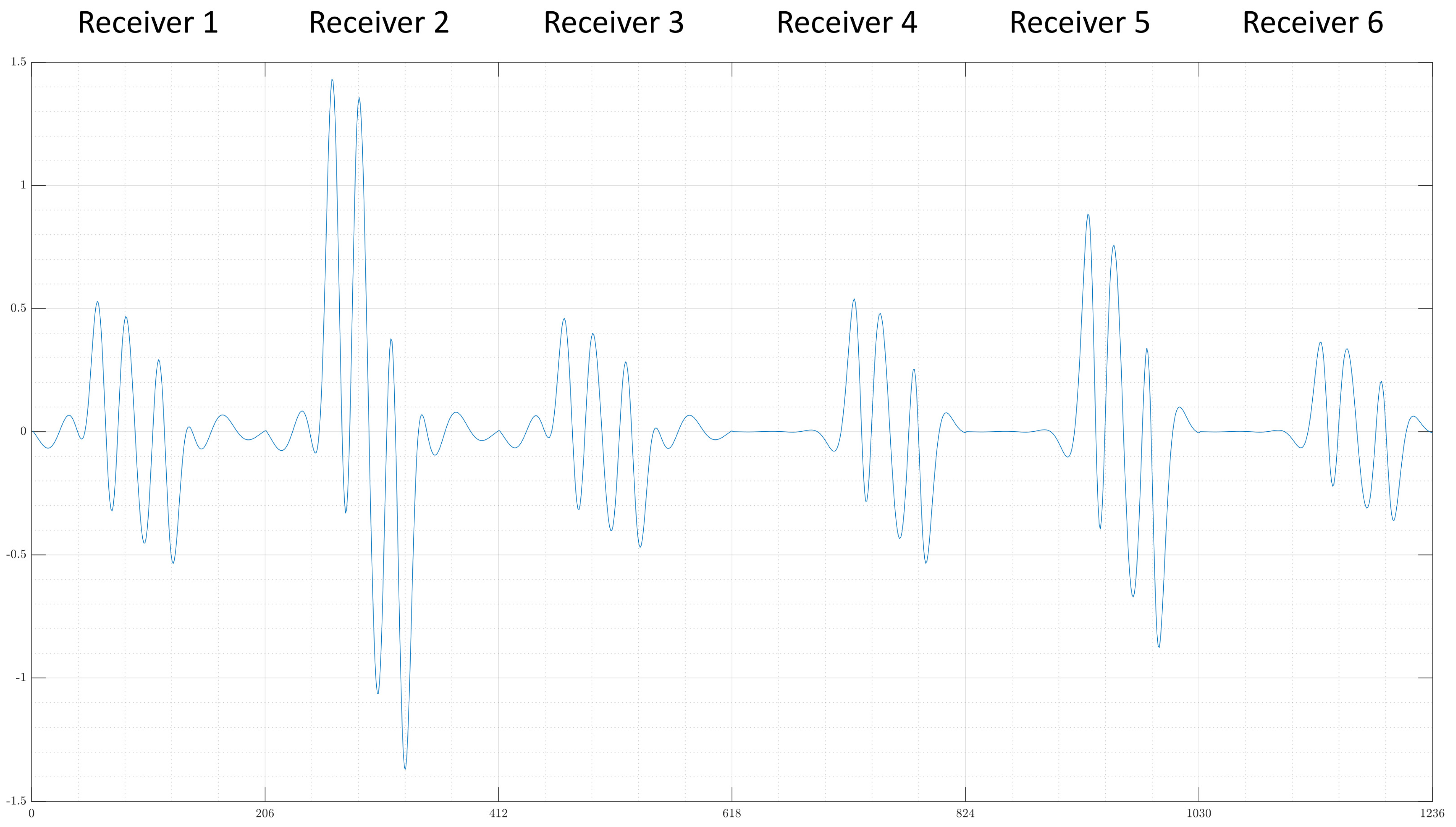
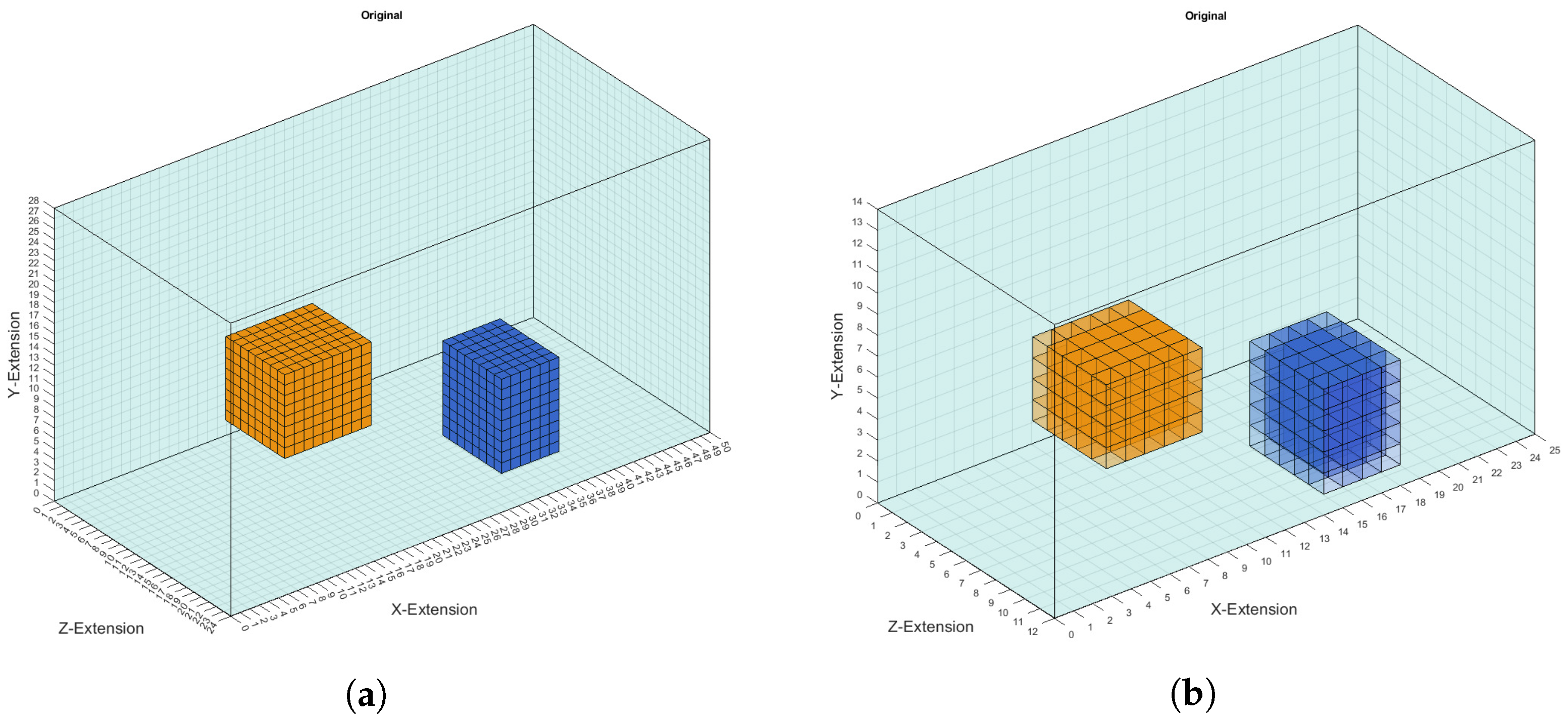
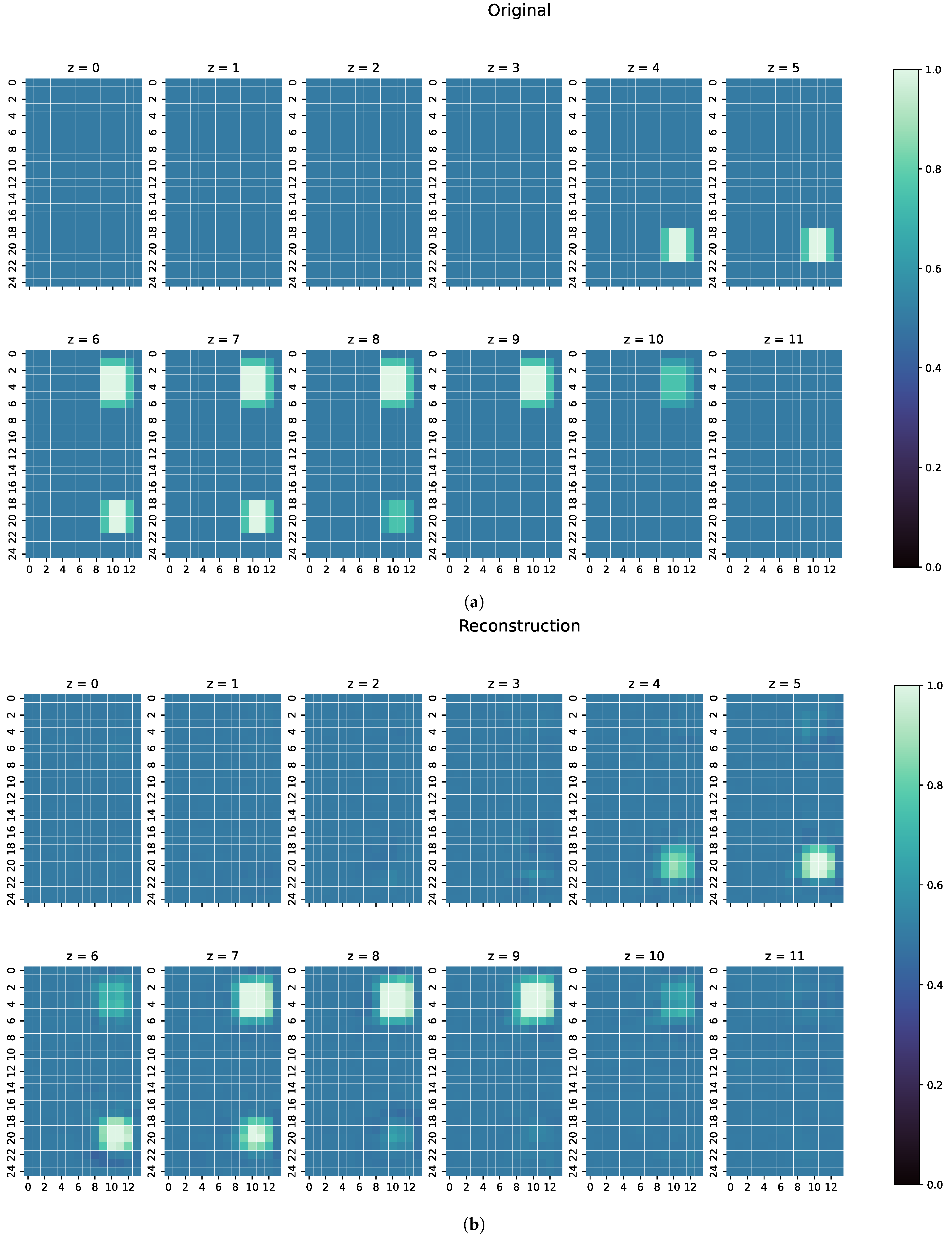
- The first examples consists of two overlapping cuboid perturbation objects, resulting in one big, noncuboid object. While generating the dataset, the perturbation objects have been ensured not to overlap, so that this case will be a completely unknown one for the network. The conductivity in this object has been set to S/m.
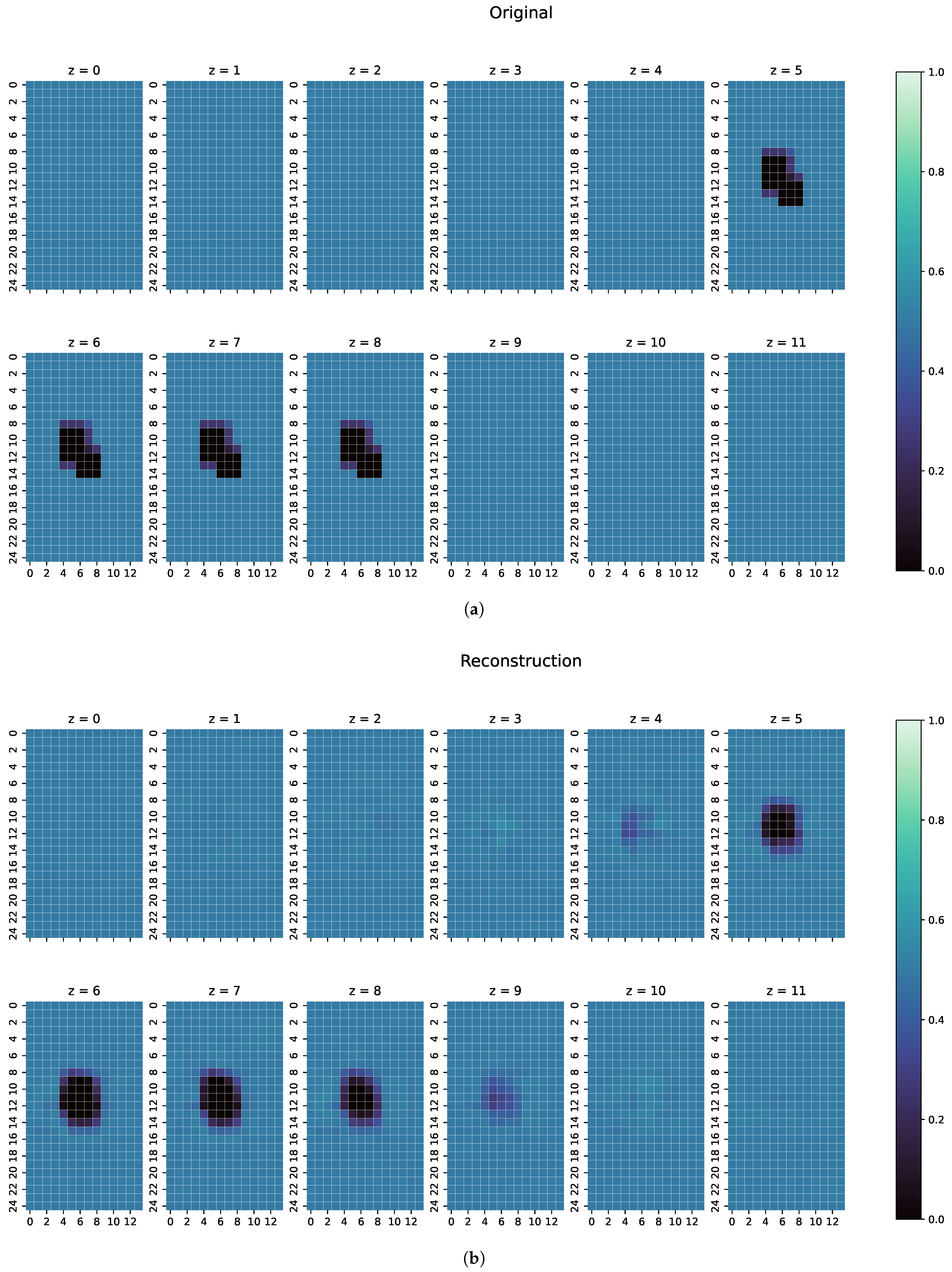
- The second example is one perturbation object of a conductivity not seen before. The training data only consists of objects with conductivity or S/m, and only on the edges of those perturbation objects do the conductivities differ from those because of the discretisation. The test case here has a conductivity of S/m.
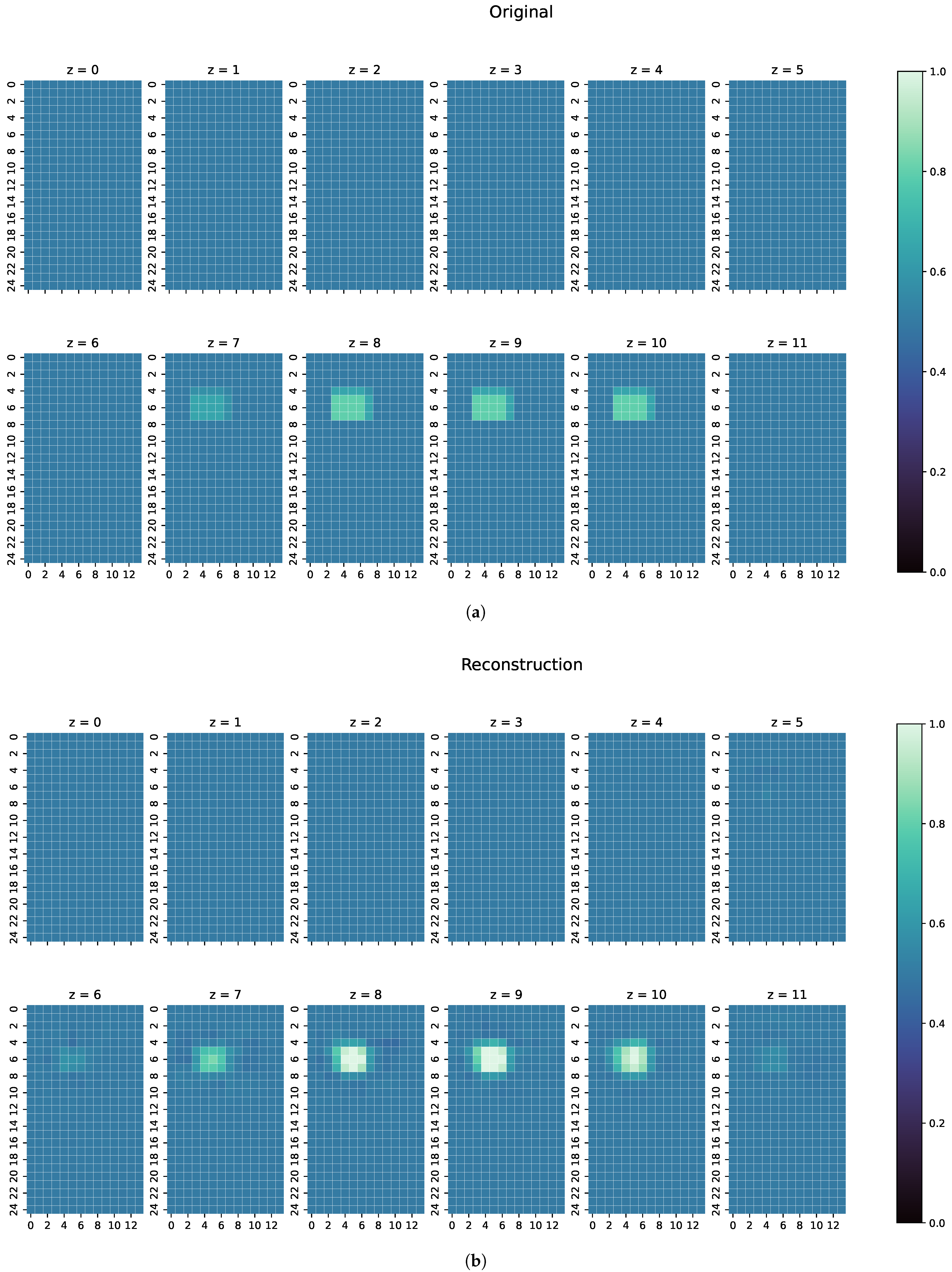
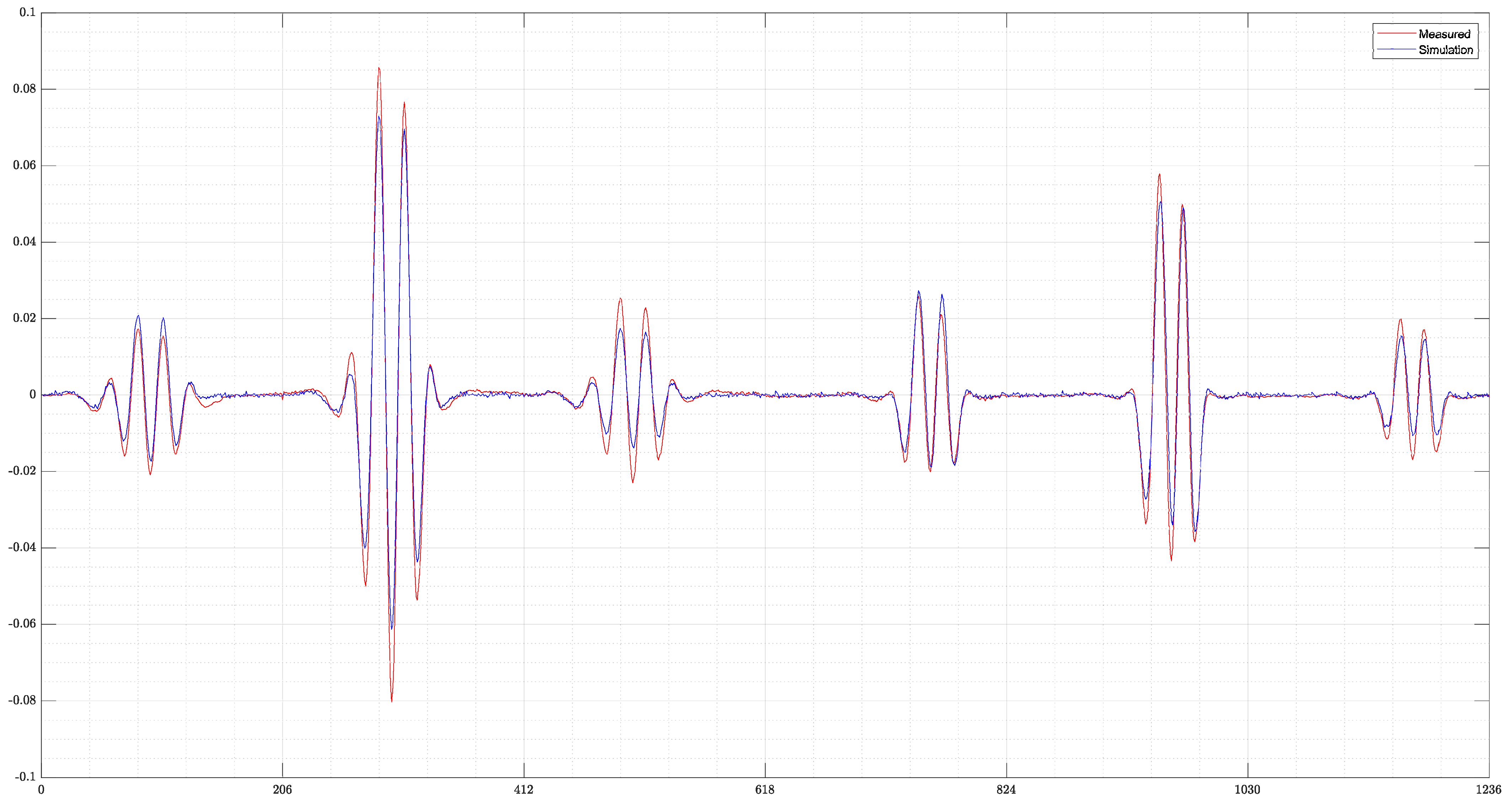
- Finally, a real measurement from the MIT-Setup is tested. In the selected measurement, the difference signals between simulation and measurement differ in amplitudes, but have a similar pattern. This shows that even the reconstruction of real measurements is possible if the pattern of the signal largely corresponds between reality and simulation.
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gürsoy, D.; Scharfetter, H. Feasibility of Lung Imaging Using Magnetic Induction Tomography. In Proceedings of the World Congress on Medical Physics and Biomedical Engineering, Munich, Germany, 7–12 September 2009. [Google Scholar] [CrossRef]
- Zolgharni, M.; Ledger, P.D.; Armitage, D.W.; Holder, D.S.; Griffiths, H. Imaging cerebral haemorrhage with magnetic induction tomography: Numerical modelling. Physiol. Meas. 2009, 30, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.-Y.; Soleimani, M. Electromagnetic Tomography for Medical and Industrial Applications: Challenges and Opportunities [Point of View]. Proc. IEEE 2013, 101, 559–565. [Google Scholar] [CrossRef]
- Klein, M.; Erni, D.; Rueter, D. Three-Dimensional Magnetic Induction Tomography: Improved Performance for the Center Regions inside a Low Conductive and Voluminous Body. Sensors 2020, 20, 1306. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Erni, D.; Rueter, D. Three-Dimensional Magnetic Induction Tomography: Practical Implementation for Imaging throughout the Depth of a Low Conductive and Voluminous Body. Sensors 2021, 21, 7725. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, M.; Lionheart, W.R.B. Image reconstruction in magnetic induction tomography using a regularized Gauss Newton method. In Proceedings of the XII International Conference on Electrical Bio-Impedance and Electrical Impedance Tomography, Gdansk, Poland, 20–24 June 2004. [Google Scholar]
- Ziolkowski, M.; Gratkowski, S.; Palka, R. Solution of Three Dimensional Inverse Problem of Magnetic Induction Tomography Using Tikhonov Regularization Method. Int. J. Appl. Electromagn. Mech. 2009, 30, 245–253. [Google Scholar] [CrossRef]
- Chen, T.; Guo, J. A computationally efficient method for sensitivity matrix calculation in magnetic induction tomography. J. Phys. Conf. Ser. 2018, 1074, 012106. [Google Scholar] [CrossRef]
- Ongie, G.; Jalal, A.; Metzler, C.A.; Baraniuk, R.G.; Dimakis, A.G.; Willett, R. Deep Learning Techniques for Inverse Problems in Imaging. IEEE J. Sel. Areas Inf. Theory 2020, 1, 39–56. [Google Scholar] [CrossRef]
- Rymarczyk, T.; Kłosowski, G.; Kozłowski, E.; Tchórzewski, P. Comparison of Selected Machine Learning Algorithms for Industrial Electrical Tomography. Sensors 2019, 19, 1521. [Google Scholar] [CrossRef] [PubMed]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. J. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef] [PubMed]
- Palka, R.; Gratkowski, S.; Baniukiewicz, P.; Komorowski, M.; Stawicki, K. Inverse Problems in Magnetic Induction Tomography of Low Conductivity Materials. Intell. Comput. Tech. Appl. Electromagn. Stud. Comput. Intell. 2008, 119, 163–170. [Google Scholar] [CrossRef]
- Yang, D.; Liu, J.; Wang, Y.; Xu, B.; Wang, X. Application of a Generative Adversarial Network in Image Reconstruction of Magnetic Induction Tomography. Sensors 2021, 21, 3869. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Huang, J.; Wang, H.; Li, B.; Zhao, Z.; Wang, J.; Wang, Y. A Novel Algorithm for High-Resolution Magnetic Induction Tomography Based on Stacked Auto-Encoder for Biological Tissue Imaging. IEEE Access 2019, 7, 185597–185606. [Google Scholar] [CrossRef]
- Kang, E.; Min, J.; Ye, J.C. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med. Phys. 2017, 44, 360–375. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.H.; Tor-Díez, C.; Meunier, H.; Bednarek, N.; Fablet, R.; Passat, N.; Rousseau, F. Multiscale brain MRI super-resolution using deep 3D convolutional networks. Comput. Med. Imaging Graph. 2019, 77, 101647. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.H.; McCann, M.T.; Froustey, E.; Unser, M. Deep Convolutional Neural Network for Inverse Problems in Imaging. IEEE Trans. Image Process. 2017, 26, 4509–4522. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef]
- Gabriel, S.; Lau, R.W.; Gabriel, C. The dielectric properties of biological tissues: {III}. Parametric models for the dielectric spectrum of tissues. IOP Publ. 1996, 41, 2271–2293. [Google Scholar] [CrossRef] [PubMed]
- Schwab, A.J. Field Theory Concepts, 1st ed.; Springer: Berlin/Heidelberg, Germany, 1988; pp. 10, 52. [Google Scholar]
- Armitage, D.W.; LeVeen, H.H.; Pethig, R. Radiofrequency-induced hyperthermia: Computer simulation of specific absorption rate distributions using realistic anatomical models. Phys Med Biol. 1983, 28, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Geselowitz, D.B. On bioelectric potentials in an inhomogeneous volume conductor. Biophys J. 1967, 7, 1–11. [Google Scholar] [CrossRef]
- Colton, D.; Kress, R. Inverse acoustic and electromagnetic scattering theory. In Applied Mathematical Sciences; Springer: Berlin, Germany, 2013; p. 154. [Google Scholar]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An introduction to statistical learning: With applications in R. In Springer Texts in Statistics; Springer: New York, NY, USA, 2013; pp. 29, 62, 217, 239. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The elements of statistical learning: Data mining, inference, and prediction. In Springer Series in Statistics; Springer: New York, NY, USA, 2001; p. 349. [Google Scholar]
- Wang, Z.; Bovik, A.C.; Sheikh H., R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
| Network | Loss | MAE | MSE | CC | SSIM |
|---|---|---|---|---|---|
| ResNet | 0.0002 | 0.0110 | 0.0013 | 0.8765 | 0.8417 |
| Example | MAE | MSE | CC | SSIM |
|---|---|---|---|---|
| Test case | 0.0092 | 0.0009 | 0.9203 | 0.8717 |
| Special case 1 | 0.0082 | 0.0008 | 0.9104 | 0.8820 |
| Special case 2 | 0.0114 | 0.0027 | 0.9396 | 0.8729 |
| Special case 3 | 0.0145 | 0.0025 | 0.7892 | 0.7802 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hofmann, A.; Klein, M.; Rueter, D.; Sauer, A. A Deep Residual Neural Network for Image Reconstruction in Biomedical 3D Magnetic Induction Tomography. Sensors 2022, 22, 7925. https://doi.org/10.3390/s22207925
Hofmann A, Klein M, Rueter D, Sauer A. A Deep Residual Neural Network for Image Reconstruction in Biomedical 3D Magnetic Induction Tomography. Sensors. 2022; 22(20):7925. https://doi.org/10.3390/s22207925
Chicago/Turabian StyleHofmann, Anna, Martin Klein, Dirk Rueter, and Andreas Sauer. 2022. "A Deep Residual Neural Network for Image Reconstruction in Biomedical 3D Magnetic Induction Tomography" Sensors 22, no. 20: 7925. https://doi.org/10.3390/s22207925
APA StyleHofmann, A., Klein, M., Rueter, D., & Sauer, A. (2022). A Deep Residual Neural Network for Image Reconstruction in Biomedical 3D Magnetic Induction Tomography. Sensors, 22(20), 7925. https://doi.org/10.3390/s22207925






