Surface Plasmon Resonance Imaging Sensor for Detection of Photolytically and Photocatalytically Degraded Glyphosate
Abstract
:1. Introduction
2. Material and Methods
2.1. Chemicals
2.2. Surface Plasmon Resonance Imaging (SPRi) and a Biochip Preparation
2.3. Total Organic Carbon (TOC) Analysis
2.4. Liquid Chromatography-Mass Spectrometry (LC–MS) Analysis
2.5. Photolytic and Photocatalytic Degradation of Glyphosate
2.6. Data Processing
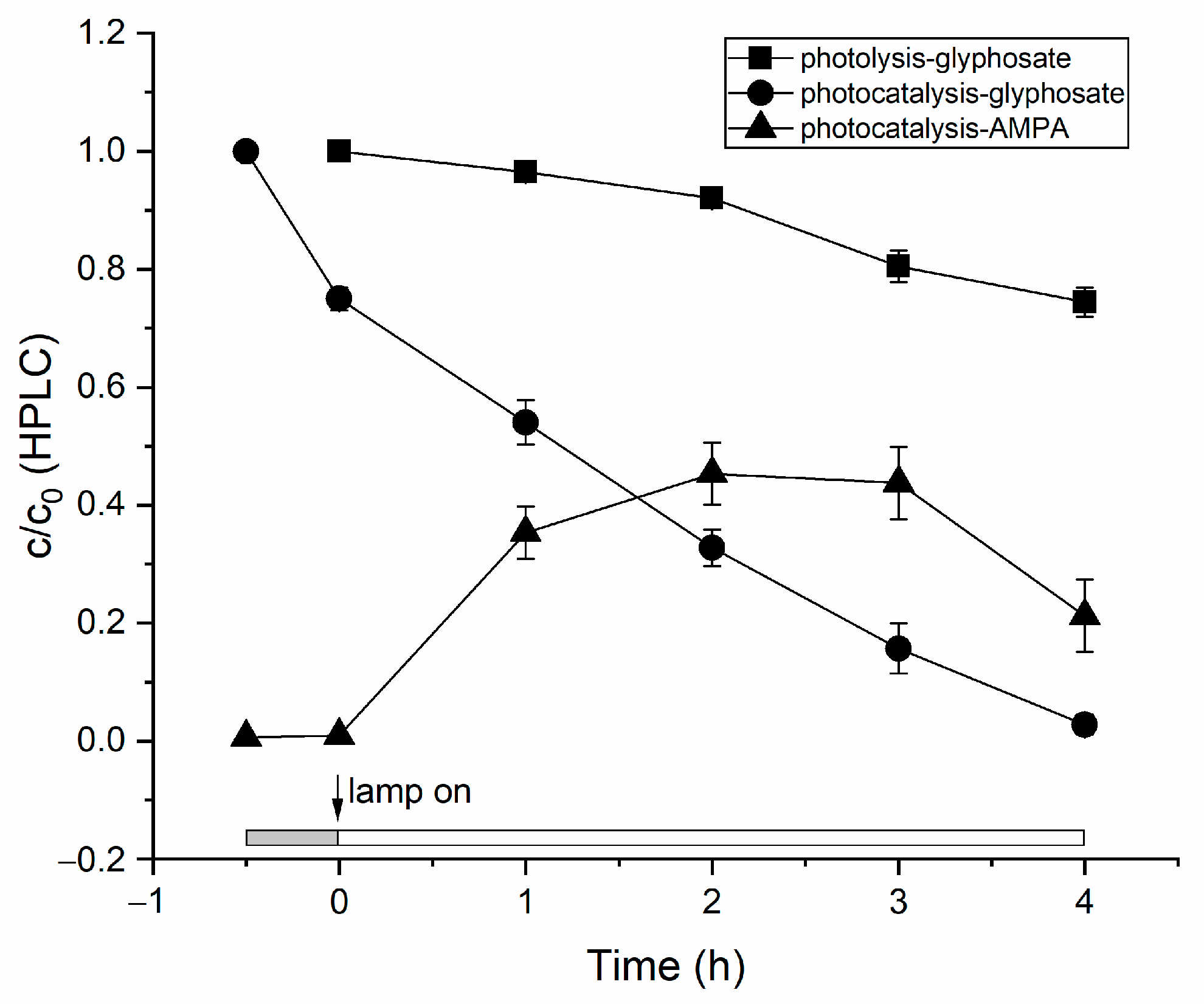
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Myers, J.P.; Antoniou, M.N.; Blumberg, B.; Carroll, L.; Colborn, T.; Everett, L.G.; Hansen, M.; Landrigan, P.J.; Lanphear, B.P.; Mesnage, R.; et al. Concerns over use of glyphosate-based herbicides and risks associated with exposures: A consensus statement. Environ. Health 2016, 15, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.W.; Smith, S.; Smith, G.; Wang, W.Q.; Li, Y.H. Glyphosate contamination in grains and foods: An overview. Food Control 2019, 106, 106710. [Google Scholar] [CrossRef]
- Guyton, K.Z.; Loomis, D.; Grosse, Y.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Scoccianti, C.; Mattock, H.; Straif, K.; International Agency for Research on Cancer Monograph Working Group. Carcinogenicity of tetrachlorvinphos, parathion, malathion, diazinon, and glyphosate. Lancet Oncol. 2015, 16, 490–491. [Google Scholar] [CrossRef]
- De Roos, A.J.; Zahm, S.H.; Cantor, K.P.; Weisenburger, D.D.; Holmes, F.F.; Burmeister, L.F.; Blair, A. Integrative assessment of multiple pesticides as risk factors for non-Hodgkin’s lymphoma among men. Occup. Environ. Med. 2003, 60, e11. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, M.; Hardell, L.; Carlberg, M.; Akerman, M. Pesticide exposure as risk factor for non-Hodgkin lymphoma including histopathological subgroup analysis. Int. J. Cancer 2008, 123, 1657–1663. [Google Scholar] [CrossRef] [PubMed]
- Feulefack, J.; Khan, A.; Forastiere, F.; Sergi, C.M. Parental Pesticide Exposure and Childhood Brain Cancer: A Systematic Review and Meta-Analysis Confirming the IARC/WHO Monographs on Some Organophosphate Insecticides and Herbicides. Child. -Basel 2021, 8, 1096. [Google Scholar] [CrossRef]
- Liao, Y.; Berthion, J.M.; Colet, I.; Merlo, M.; Nougadere, A.; Hu, R.W. Validation and application of analytical method for glyphosate and glufosinate in foods by liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2018, 1549, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Martins, H.A.; Lebre, D.T.; Wang, A.Y.; Pires, M.A.F.; Bustillos, O.V. An alternative and fast method for determination of glyphosate and aminomethylphosphonic acid (AMPA) residues in soybean using liquid chromatography coupled with tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2009, 23, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.D.; Feng, Y.R.; Ma, L.; An, J.; Zhang, H.Y.; Cao, M.S.; Zhu, H.J.; Kang, W.J.; Lian, K.Q. A method for determining glyphosate and its metabolite aminomethyl phosphonic acid by gas chromatography-flame photometric detection. J. Chromatogr. A 2019, 1589, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Aydin, Z.; Keles, M. A reaction-based system for the colorimetric detection of glyphosate in real samples. Spectrochim. Acta Part A-Mol. Biomol. Spectrosc. 2022, 267, 120501. [Google Scholar] [CrossRef] [PubMed]
- De Goes, R.E.; Muller, M.; Fabris, J.L. Spectroscopic Detection of Glyphosate in Water Assisted by Laser-Ablated Silver Nanoparticles. Sensors 2017, 17, 954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.L.; Gao, Y.; Li, Y.L.; Li, X.L.; Zhang, H.J.; Han, X.X.; Zhao, B.; Su, L. Indirect glyphosate detection based on ninhydrin reaction and surface-enhanced Raman scattering spectroscopy. Spectrochim. Acta Part A-Mol. Biomol. Spectrosc. 2018, 197, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Crocoli, L.C.; Ortiz, R.S.; Moura, S. Development and validation of a qNMR method for analyses of legal and illegal formulations of glyphosate. Anal. Methods 2019, 11, 4052–4059. [Google Scholar] [CrossRef]
- Byer, J.D.; Struger, J.; Klawunn, P.; Todd, A.; Sverko, E. Low cost monitoring of glyphosate in surface waters using the ELISA method: An evaluation. Environ. Sci. Technol. 2008, 42, 6052–6057. [Google Scholar] [CrossRef]
- Rubio, F.; Veldhuis, L.J.; Clegg, B.S.; Fleeker, J.R.; Hall, J.C. Comparison of a direct ELISA and an HPLC method for glyphosate determinations in water. J. Agric. Food Chem. 2003, 51, 691–696. [Google Scholar] [CrossRef]
- See, H.H.; Hauser, P.C.; Ibrahim, W.A.W.; Sanagi, M.M. Rapid and direct determination of glyphosate, glufosinate, and aminophosphonic acid by online preconcentration CE with contactless conductivity detection. Electrophoresis 2010, 31, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Csapó, E.; Juhász, Á.; Varga, N.; Sebők, D.; Hornok, V.; Janovák, L.; Dékány, I. Thermodynamic and kinetic characterization of pH-dependent interactions between bovine serum albumin and ibuprofen in 2D and 3D systems. Colloids Surf. A Physicochem. Eng. Asp. 2016, 504, 471–478. [Google Scholar] [CrossRef] [Green Version]
- Homola, J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef] [PubMed]
- Hornok, V.; Juhasz, A.; Paragi, G.; Kovacs, A.N.; Csapo, E. Thermodynamic and kinetic insights into the interaction of kynurenic acid with human serum albumin: Spectroscopic and calorimetric approaches. J. Mol. Liq. 2020, 313, 112869. [Google Scholar] [CrossRef]
- Ding, X.K.; Yang, K.L. Development of an Oligopeptide Functionalized Surface Plasmon Resonance Biosensor for Online Detection of Glyphosate. Anal. Chem. 2013, 85, 5727–5733. [Google Scholar] [CrossRef]
- Do, M.H.; Dubreuil, B.; Peydecastaing, J.M.; Vaca-Medina, G.; Nhu-Trang, T.T.; Jaffrezic-Renault, N.; Behra, P. Chitosan-Based Nanocomposites for Glyphosate Detection Using Surface Plasmon Resonance Sensor. Sensors 2020, 20, 5942. [Google Scholar] [CrossRef]
- Feng, D.; Soric, A.; Boutin, O. Treatment technologies and degradation pathways of glyphosate: A critical review. Sci. Total Environ. 2020, 742, 140559. [Google Scholar] [CrossRef] [PubMed]
- Dosnon-Olette, R.; Couderchet, M.; Oturan, M.A.; Oturan, N.; Eullaffroy, P. Potential use of lemna minor for the phytoremediation of isoproturon and glyphosate. Int. J. Phytoremediation 2011, 13, 601–612. [Google Scholar] [CrossRef]
- Zhan, H.; Feng, Y.M.; Fan, X.H.; Chen, S.H. Recent advances in glyphosate biodegradation. Appl. Microbiol. Biotechnol. 2018, 102, 5033–5043. [Google Scholar] [CrossRef] [PubMed]
- Vera, M.S.; Lagomarsino, L.; Sylvester, M.; Perez, G.L.; Rodriguez, P.; Mugni, H.; Sinistro, R.; Ferraro, M.; Bonetto, C.; Zagarese, H.; et al. New evidences of Roundup(A (R)) (glyphosate formulation) impact on the periphyton community and the water quality of freshwater ecosystems. Ecotoxicology 2010, 19, 710–721. [Google Scholar] [CrossRef]
- Song, J.F.; Li, X.M.; Figoli, A.; Huang, H.; Pan, C.; He, T.; Jiang, B. Composite hollow fiber nanofiltration membranes for recovery of glyphosate from saline wastewater. Water Res. 2013, 47, 2065–2074. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Duan, J.M.; Saint, C.P.; Mulcahy, D. Removal of glyphosate and aminomethylphosphonic acid from synthetic water by nanofiltration. Environ. Technol. 2018, 39, 1384–1392. [Google Scholar] [CrossRef] [PubMed]
- Pereira, H.A.; Hernandes, P.R.T.; Netto, M.S.; Reske, G.D.; Vieceli, V.; Oliveira, L.F.S.; Dotto, G.L. Adsorbents for glyphosate removal in contaminated waters: A review. Environ. Chem. Lett. 2021, 19, 1525–1543. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.F.; Yang, Q.Z.; Evans, D.G.; Forano, C.; Duan, X. Study on adsorption of glyphosate (N-phosphonomethyl glycine) pesticide on MgAl-layered double hydroxides in aqueous solution. J. Hazard. Mater. 2005, 125, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Manassero, A.; Passalia, C.; Negro, A.C.; Cassano, A.E.; Zalazar, C.S. Glyphosate degradation in water employing the H2O2/UVC process. Water Res. 2010, 44, 3875–3882. [Google Scholar] [CrossRef] [PubMed]
- Balci, B.; Oturan, M.A.; Oturan, N.; Sires, I. Decontamination of Aqueous Glyphosate, (Aminomethyl) phosphonic Acid, and Glufosinate Solutions by Electro-Fenton-like Process with Mn2+ as the Catalyst. J. Agric. Food Chem. 2009, 57, 4888–4894. [Google Scholar] [CrossRef]
- Echavia, G.R.M.; Matzusawa, F.; Negishi, N. Photocatalytic degradation of organophosphate and phosphonoglycine pesticides using TiO2 immobilized on silica gel. Chemosphere 2009, 76, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Rubi-Juarez, H.; Cotillas, S.; Saez, C.; Canizares, P.; Barrera-Diaz, C.; Rodrigo, M.A. Removal of herbicide glyphosate by conductive-diamond electrochemical oxidation. Appl. Catal. B-Environ. 2016, 188, 305–312. [Google Scholar] [CrossRef]
- Samsel, A.; Seneff, S. Glyphosate’s Suppression of Cytochrome P450 Enzymes and Amino Acid Biosynthesis by the Gut Microbiome: Pathways to Modern Diseases. Entropy 2013, 15, 1416–1463. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface Plasmon Resonance: A Versatile Technique for Biosensor Applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Chen, Y.F.; Wang, H.; Hu, S.Q.; Xia, K.; Xiong, X.; Huang, W.J.; Lu, H.H.; Yu, J.H.; Guan, H.Y.; et al. Titanium dioxide nanoparticle modified plasmonic interface for enhanced refractometric and biomolecular sensing. Opt. Express 2018, 26, 33226–33237. [Google Scholar] [CrossRef]
- Scarano, S.; Mascini, M.; Turner, A.P.F.; Minunni, M. Surface plasmon resonance imaging for affinity-based biosensors. Biosens. Bioelectron. 2010, 25, 957–966. [Google Scholar] [CrossRef] [Green Version]
- Nebert, D.W.; Wikvall, K.; Miller, W.L. Human cytochromes P450 in health and disease. Philos. Trans. R. Soc. B-Biol. Sci. 2013, 368, 20120431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karamanska, R.; Clarke, J.; Blixt, O.; MacRae, J.I.; Zhang, J.Q.Q.; Crocker, P.R.; Laurent, N.; Wright, A.; Flitsch, S.L.; Russell, D.A.; et al. Surface plasmon resonance imaging for real-time, label-free analysis of protein interactions with carbohydrate microarrays. Glycoconj. J. 2008, 25, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Papagiannaki, D.; Medana, C.; Binetti, R.; Calza, P.; Roslev, P. Effect of UV-A, UV-B and UV-C irradiation of glyphosate on photolysis and mitigation of aquatic toxicity. Sci. Rep. 2020, 10, 20247. [Google Scholar] [CrossRef] [PubMed]
- Law, W.-C.; Yong, K.-T.; Baev, A.; Hu, R.; Prasad, P.N. Nanoparticle enhanced surface plasmon resonance biosensing: Application of gold nanorods. Opt. Express 2009, 17, 19041–19046. [Google Scholar] [CrossRef]
- Park, J.-H.; Cho, Y.-W.; Kim, T.-H. Recent Advances in Surface Plasmon Resonance Sensors for Sensitive Optical Detection of Pathogens. Biosensors 2022, 12, 180. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.Q.; Yu, F.; Zare, R.N. Microfluidic device for immunoassays based on surface plasmon resonance imaging. Lab A Chip 2008, 8, 694–700. [Google Scholar] [CrossRef]
- Nafisah, S.; Morsin, M.; Jumadi, N.A.; Nayan, N.; Shah, N.S.M.; Razali, N.L.; An’Nisa, N.Z. Improved Sensitivity and Selectivity of Direct Localized Surface Plasmon Resonance Sensor Using Gold Nanobipyramids for Glyphosate Detection. Ieee Sens. J. 2020, 20, 2378–2389. [Google Scholar] [CrossRef]
- Li, M.C.; Chen, K.R.; Kuo, C.C.; Lin, Y.X.; Su, L.C. A Simple Phase-Sensitive Surface Plasmon Resonance Sensor Based on Simultaneous Polarization Measurement Strategy. Sensors 2021, 21, 7615. [Google Scholar] [CrossRef] [PubMed]
- Balciunas, D.; Plausinaitis, D.; Ratautaite, V.; Ramanaviciene, A.; Ramanavicius, A. Towards electrochemical surface plasmon resonance sensor based on the molecularly imprinted polypyrrole for glyphosate sensing. Talanta 2022, 241, 123252. [Google Scholar] [CrossRef]
- Steiner, G. Surface plasmon resonance imaging. Anal. Bioanal. Chem. 2004, 379, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, E.; Lausted, C.; Lin, T.; Yang, C.W.T.; Hood, L.; Lagally, E.T. Parallel microfluidic surface plasmon resonance imaging arrays. Lab A Chip 2010, 10, 581–588. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vráblová, M.; Smutná, K.; Koutník, I.; Prostějovský, T.; Žebrák, R. Surface Plasmon Resonance Imaging Sensor for Detection of Photolytically and Photocatalytically Degraded Glyphosate. Sensors 2022, 22, 9217. https://doi.org/10.3390/s22239217
Vráblová M, Smutná K, Koutník I, Prostějovský T, Žebrák R. Surface Plasmon Resonance Imaging Sensor for Detection of Photolytically and Photocatalytically Degraded Glyphosate. Sensors. 2022; 22(23):9217. https://doi.org/10.3390/s22239217
Chicago/Turabian StyleVráblová, Martina, Kateřina Smutná, Ivan Koutník, Tomáš Prostějovský, and Radim Žebrák. 2022. "Surface Plasmon Resonance Imaging Sensor for Detection of Photolytically and Photocatalytically Degraded Glyphosate" Sensors 22, no. 23: 9217. https://doi.org/10.3390/s22239217
APA StyleVráblová, M., Smutná, K., Koutník, I., Prostějovský, T., & Žebrák, R. (2022). Surface Plasmon Resonance Imaging Sensor for Detection of Photolytically and Photocatalytically Degraded Glyphosate. Sensors, 22(23), 9217. https://doi.org/10.3390/s22239217





