Multi-Layer Graph Attention Network for Sleep Stage Classification Based on EEG
Abstract
1. Introduction
2. Related Work
3. Materials and Methods
3.1. Preliminaries
3.2. Spatial Node-Level Graph Attention Sleep Network
3.2.1. Multi-Head Graph Attention Learning
3.2.2. Spatial Domain Attention
3.3. Temporal Node-Level Graph Attention Sleep Network
3.3.1. Time-Frequency Feature Extraction
3.3.2. Temporal Domain Attention
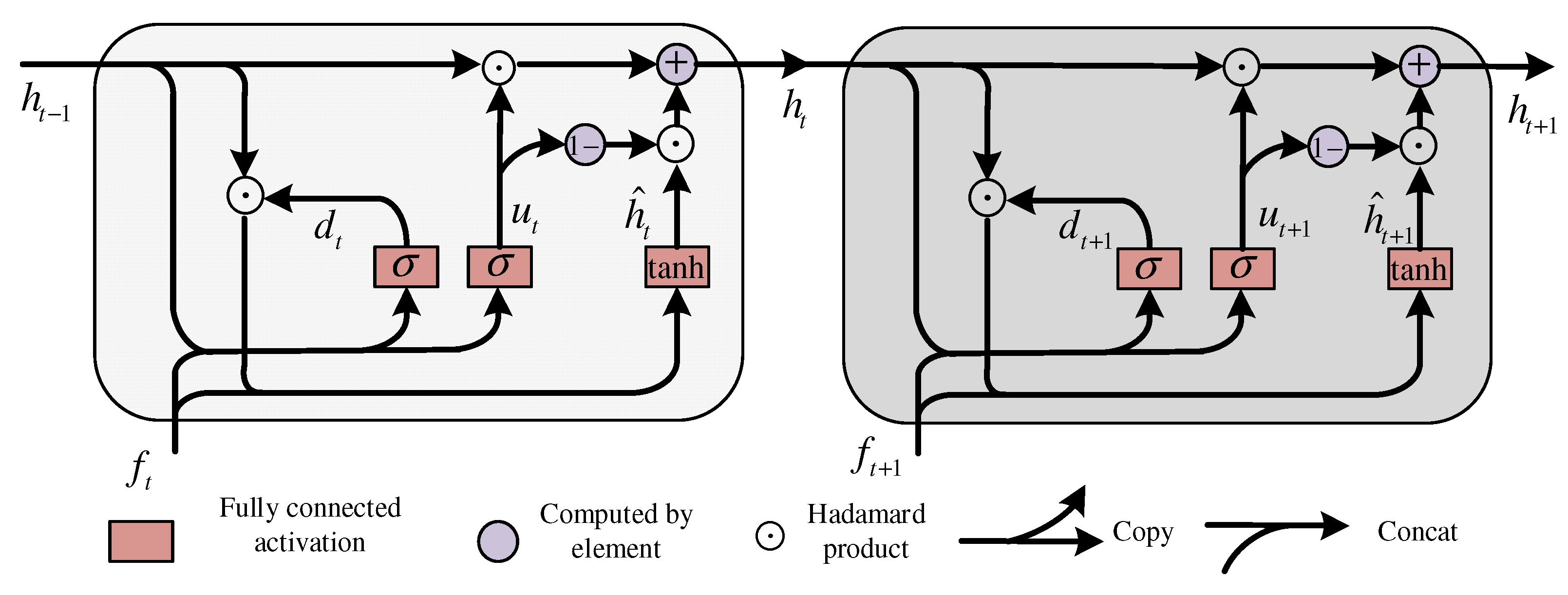
3.4. Stage-Level Attention Transitional Estimator
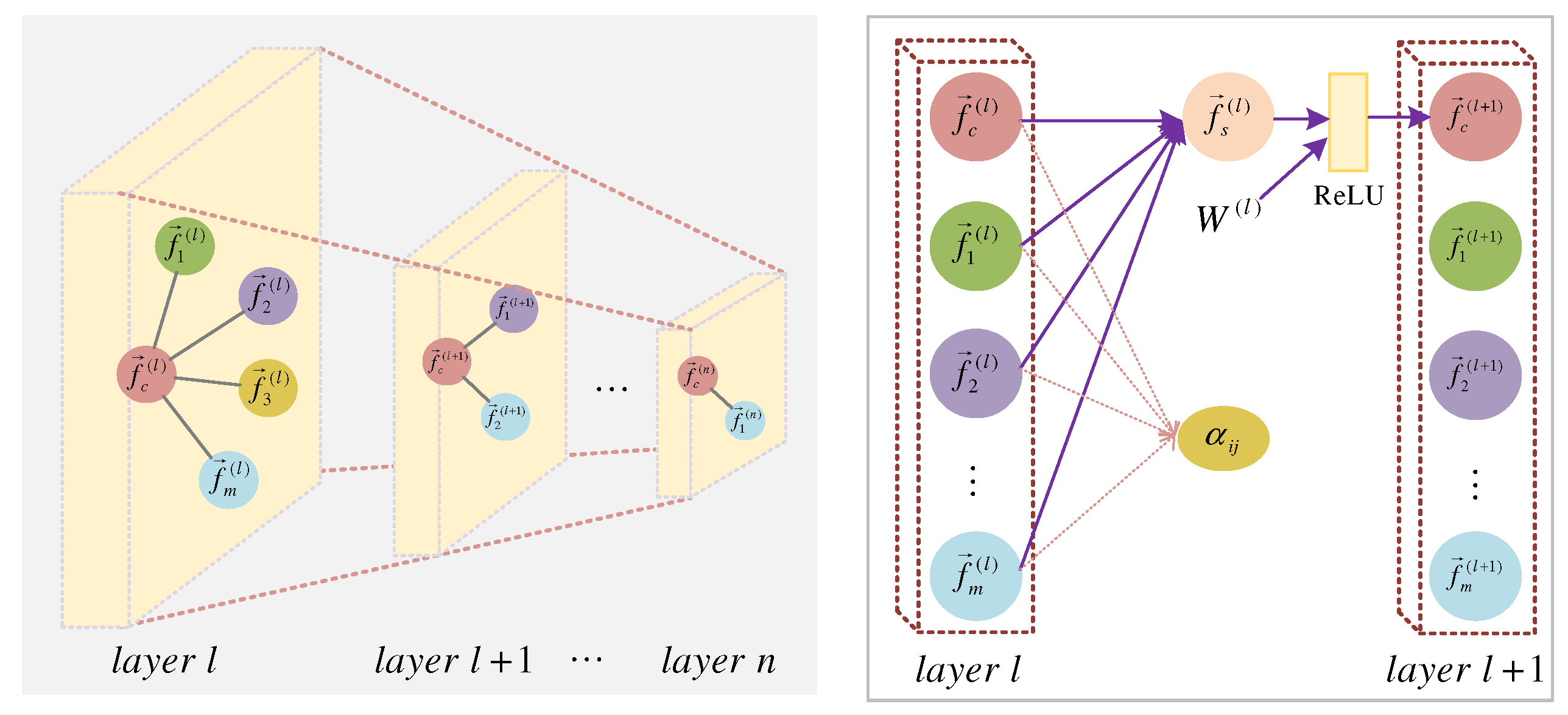
3.5. Spatial Multi-Layer Graph Attention Convolution
4. Results and Discussion
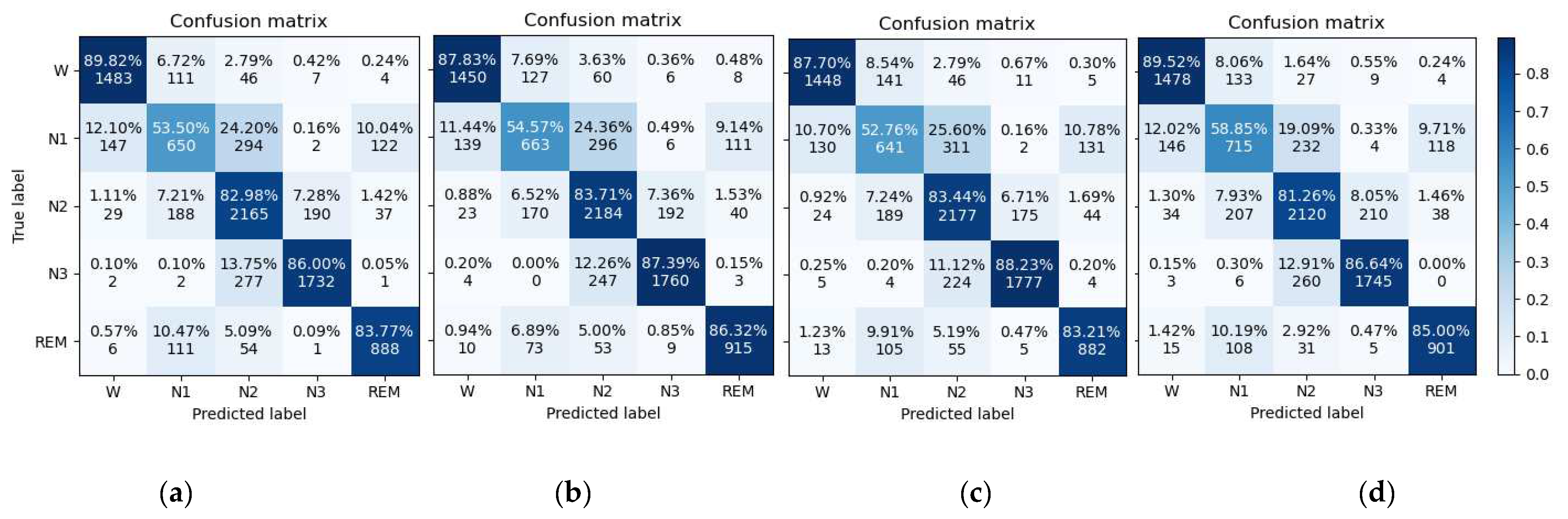
4.1. Classification Performance
4.2. Number of Parameters and Training Duration
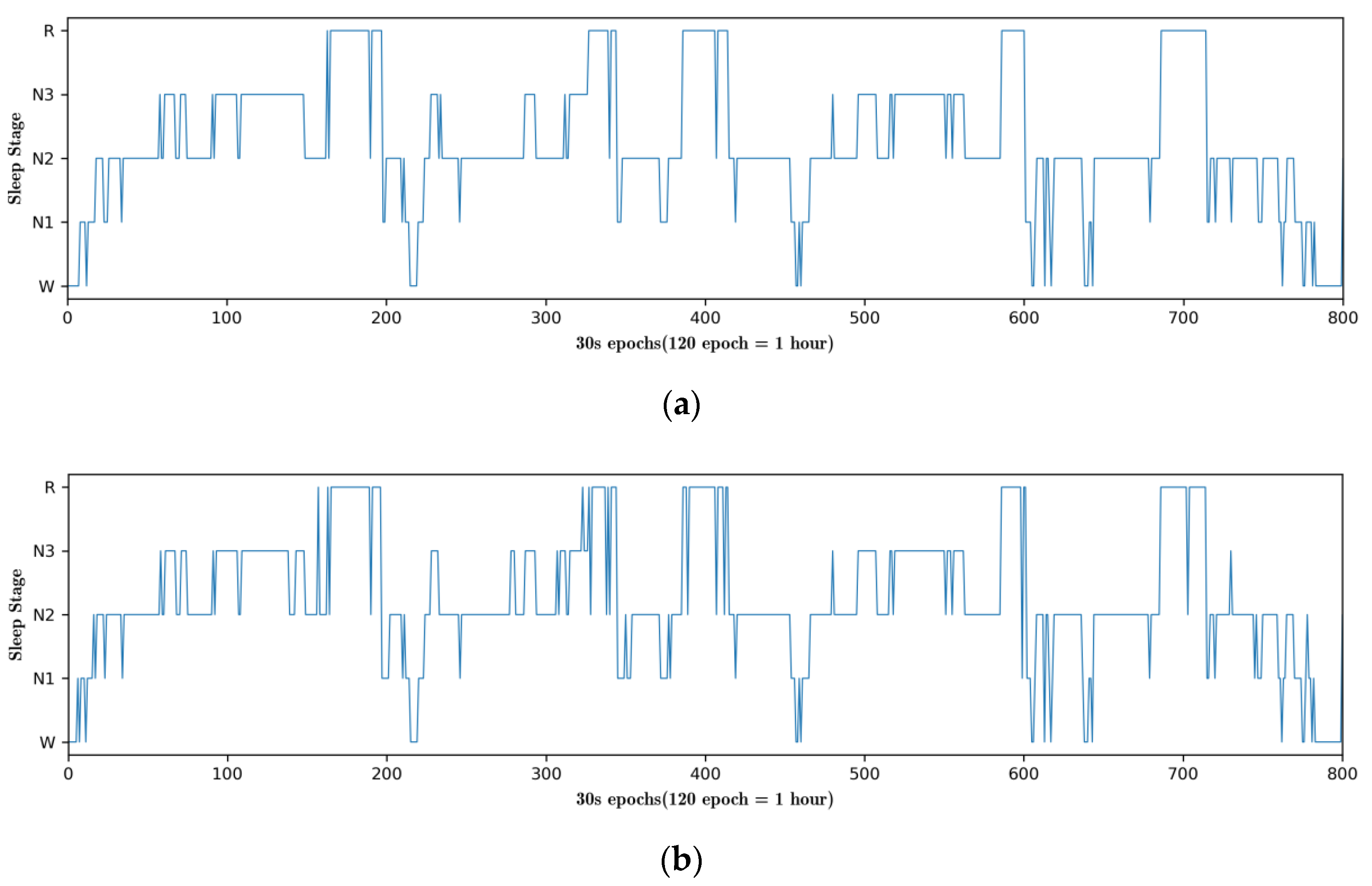
4.3. Hypnograms
4.4. Ablation Experiments Results
4.5. Heads Numbers
4.6. Speed Training
4.7. Attention Channel Visualization
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Dataset and Experiment Settings
| Parameter | Batch Size | Learning Rate | Reduction Rate | Optimizer | Epochs | |||
|---|---|---|---|---|---|---|---|---|
| Value | 32 | 0.0002 | 2 | Adam | 80 | 0.9 | 0.999 |
Appendix A.2. Baseline
Appendix A.3. Pseudocode
| Algorithm A1: MGANet for sleep staging |
| Input: An information network: |
| Output: Node vector fusion information representations |
| Initialization: , training epochs I and labeled nodes |
| While do |
| for a vertex do |
| //Learn sleep vector for each spatial feature. |
| //Learn attention spatial feature representation. |
| //Learn the sleep vector for each temporal feature by Equation (8). |
| //Learn attention temporal feature representation. |
| end |
| //Calculate attention coefficient. |
| //sleep stage probability distribution. |
| //epoch features and transitional fusion information. |
| end |
References
- Carskadon, M.A.; Rechtschaffen, A. Monitoring and staging human sleep. Princ. Pract. Sleep Med. 2011, 5, 16–26. [Google Scholar]
- Killgore, W. Effects of sleep deprivation on cognition. Prog. Brain Res. 2010, 185, 105–129. [Google Scholar] [PubMed]
- Acharya, R.; Faust, O.; Kannathal, N.; Chua, T.; Laxminarayan, S. Non-linear analysis of EEG signals at various sleep stages. Comput. Methods Programs Biomed. 2005, 80, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Berry, B.; Brooks, R.; Gamaldo, C. The AASM manual for the scoring of sleep and associated events. In Rules, Terminology and Technical Specifications; American Academy of Sleep Medicine: Darien, IL, USA, 2012; Volume 176, p. 2012. [Google Scholar]
- Rechtschaffen, A. A manual for standardized terminology, techniques and scoring system for sleep stages in human subjects. In Brain Information Service; Brain Research Institute, US Department of Health, Education and Welfare: Potomac, MD, USA, 1968. [Google Scholar]
- Berry, R.; Budhiraja, R.; Gottlieb, D.J. Rules for scoring respiratory events in sleep: Update of the 2007 AASM manual for the scoring of sleep and associated events: Deliberations of the sleep apnea definitions task force of the American Academy of Sleep Medicine. J. Clin. Sleep Med. 2012, 8, 597–619. [Google Scholar] [CrossRef]
- Ghimatgar, H.; Kazemi, K.; Helfroush, M.S.; Aarabi, A. An automatic single-channel EEG-based sleep stage scoring method based on hidden Markov Model. J. Neurosci. Methods 2019, 41, 108320. [Google Scholar] [CrossRef]
- Memar, P.; Faradji, F. A novel multi-class EEG-based sleep stage classifification system. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 26, 84–95. [Google Scholar] [CrossRef]
- Alickovic, E.; Subasi, A. Ensemble SVM method for automatic sleep stage classification. IEEE Trans. Instrum. Meas. 2018, 67, 1258–1265. [Google Scholar] [CrossRef]
- Supratak, A.; Dong, H.; Wu, C. DeepSleepNet: A model for automatic sleep stage scoring based on raw single-channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1998–2008. [Google Scholar] [CrossRef]
- Chambon, S.; Galtier, M.N.; Arnal, P.J.; Wainrib, G.; Gramfort, A. A deep learning architecture for temporal sleep stage classifification using multivariate and multimodal time series. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 758–769. [Google Scholar] [CrossRef]
- Dong, H.; Supratak, A.; Pan, W. Mixed neural network approach for temporal sleep stage classification. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 26, 324–333. [Google Scholar] [CrossRef]
- Tsinalis, O.; Matthews, P.M.; Guo, Y. Automatic sleep stage scoring using time-frequency analysis and stacked sparse autoencoders. Ann. Biomed. Eng. 2016, 44, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Sekkal, R.N.; Bereksi-Reguig, F.; Ruiz-Fernandez, D. Automatic sleep stage classification: From classical machine learning methods to deep learning. Biomed. Signal Process. Control 2022, 77, 103751. [Google Scholar] [CrossRef]
- Honey, C.J.; Thivierge, J.P.; Sporns, O. Can structure predict function in the human brain? Neuroimage 2010, 52, 766–776. [Google Scholar] [CrossRef]
- Jia, Z.; Lin, Y.; Wang, J. GraphSleepNet: Adaptive Spatial-Temporal Graph Convolutional Networks for Sleep Stage Classification. In Proceedings of the Twenty-Ninth International Joint Conference on Artificial Intelligence, Yokohama, Japan, 11–17 July 2020; pp. 1324–1330. [Google Scholar]
- Liang, Z.; Zhou, R.; Zhang, L. EEGFuseNet: Hybrid unsupervised deep feature characterization and fusion for high-dimensional EEG with an application to emotion recognition. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 1913–1925. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.; Olsen, L.; Lindow, M. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS ONE 2007, 2, e873. [Google Scholar] [CrossRef] [PubMed]
- Veličković, P.; Cucurull, G.; Casanova, A. Graph Attention Networks. Int. Conf. Learn. Represent. 2018, 1050, 4. [Google Scholar]
- Allam, J.P.; Samantray, S.; Behara, C. Customized deep learning algorithm for drowsiness detection using single-channel EEG signal. In Artificial Intelligence-Based Brain-Computer Interface; Academic Press: Cambridge, MA, USA, 2022; pp. 189–201. [Google Scholar]
- Bik, A.; Sam, C.; de Groot, E.R. A scoping review of behavioral sleep stage classification methods for preterm infants. Sleep Med. 2022, 90, 74–82. [Google Scholar] [CrossRef]
- Phan, H.; Andreotti, F.; Cooray, N. SeqSleepNet: End-to-end hierarchical recurrent neural network for sequence-to-sequence automatic sleep staging. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 400–410. [Google Scholar] [CrossRef]
- Chriskos, P.; Frantzidis, C.A.; Gkivogkli, P.T. Automatic sleep staging employing convolutional neural networks and cortical connectivity images. IEEE Trans. Neural Netw. Learn. Syst. 2019, 31, 113–123. [Google Scholar] [CrossRef]
- Jia, Z.; Cai, X.; Zheng, G. SleepPrintNet: A multivariate multimodal neural network based on physiological time-series for automatic sleep staging. IEEE Trans. Artif. Intell. 2020, 1, 248–257. [Google Scholar] [CrossRef]
- Eldele, E.; Chen, Z.; Liu, C. An attention-based deep learning approach for sleep stage classification with single-channel eeg. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Alexander, N.O. Automatic sleep stage classifification with deep residual networks in a mixed-cohort setting. Sleep 2021, 44, 161. [Google Scholar]
- Jia, Z.; Lin, Y.; Wang, J. Multi-view spatial-temporal graph convolutional networks with domain generalization for sleep stage classification. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 1977–1986. [Google Scholar] [CrossRef]
- Khare, S.K.; Bajaj, V. Time–frequency representation and convolutional neural network-based emotion recognition. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 2901–2909. [Google Scholar] [CrossRef]
- Arias-Vergara, T.; Klumpp, P.; Vasquez-Correa, J.C.; Nöth, E.; Orozco-Arroyave, J.R.; Schuster, M. Multi-channel spectrograms for speech processing applications using deep learning methods. Pattern Anal. Appl. 2021, 24, 423–431. [Google Scholar] [CrossRef]
- Lopac, N.; Hržić, F.; Vuksanović, I.P.; Lerga, J. Detection of Non-Stationary GW Signals in High Noise From Cohen’s Class of Time–Frequency Representations Using Deep Learning. IEEE Access 2021, 10, 2408–2428. [Google Scholar] [CrossRef]
- Khalighi, S.; Sousa, T.; Santos, J.M. ISRUC-Sleep: A comprehensive public dataset for sleep researchers. Comput. Methods Programs Biomed. 2016, 124, 180–192. [Google Scholar] [CrossRef]
- Guillot, A.; Sauvet, F.; During, E.H. Dreem open datasets: Multi-scored sleep datasets to compare human and automated sleep staging. IEEE Trans. Neural Syst. Rehabil. Eng. 2020, 28, 1955–1965. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Cui, L.; Mueller, R. The National Sleep Research Resource: Towards a sleep data commons. J. Am. Med. Inform. Assoc. 2018, 25, 1351–1358. [Google Scholar] [CrossRef]
- Quan, S.F.; Howard, B.V.; Iber, C.; Kiley, J.P.; Nieto, F.J.; O’Connor, G.T.; Rapoport, D.M.; Redline, S.; Robbins, J.; Samet, J.M.; et al. The sleep heart health study: Design, rationale, and methods. Sleep 1997, 20, 1077–1085. [Google Scholar]
- Foulkes, W.D. Dream reports from different stages of sleep. J. Abnorm. Soc. Psychol. 1962, 65, 14. [Google Scholar] [CrossRef]
- Carskadon, M.A.; Dement, W.C. Normal human sleep: An overview. Princ. Pract. Sleep Med. 2005, 4, 13–23. [Google Scholar]
- Batista, G.E.; Prati, R.C.; Monard, M.C. A study of the behavior of several methods for balancing machine learning training data. ACM SIGKDD Explor. Newsl. 2004, 6, 20–29. [Google Scholar] [CrossRef]





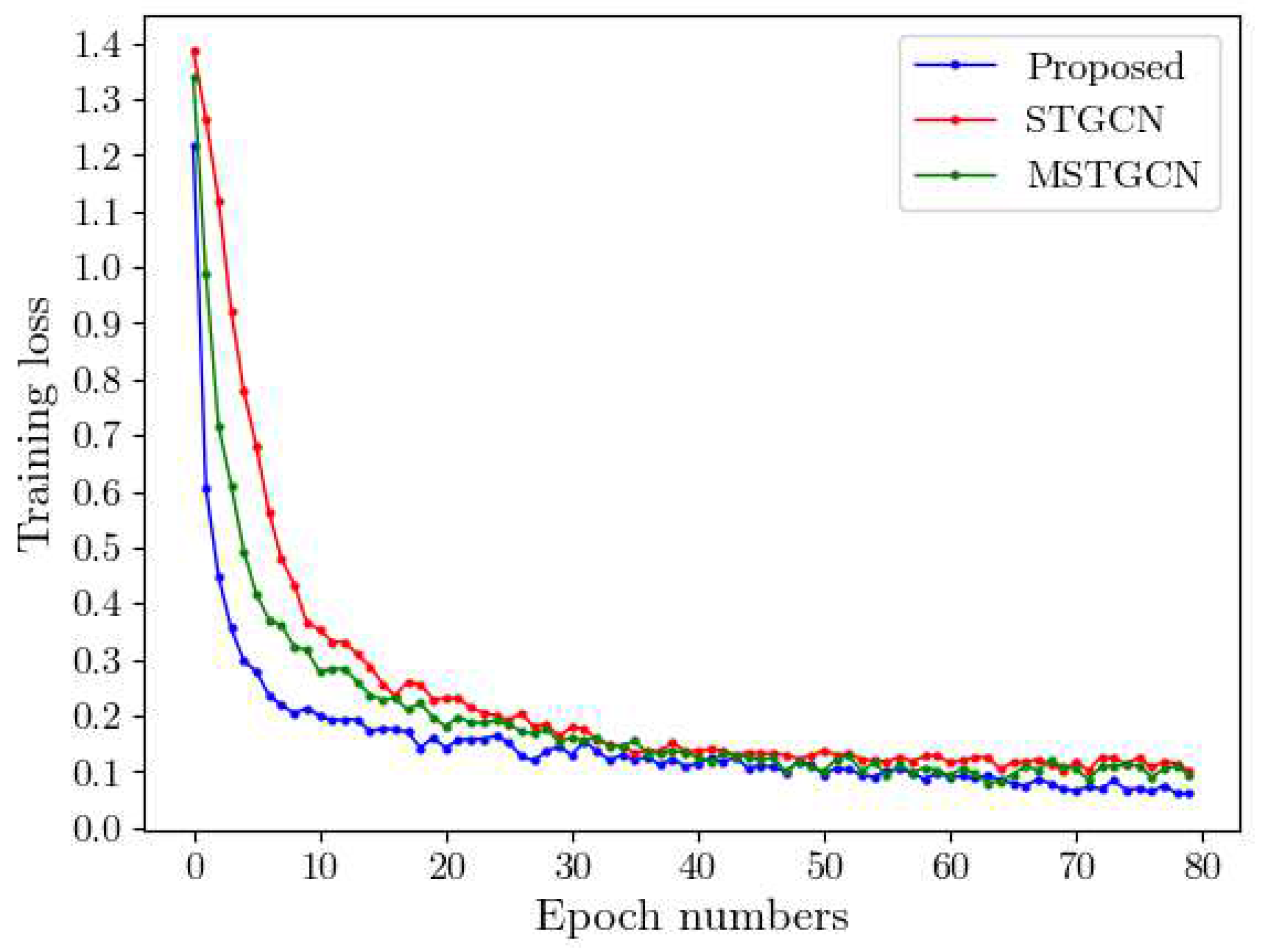
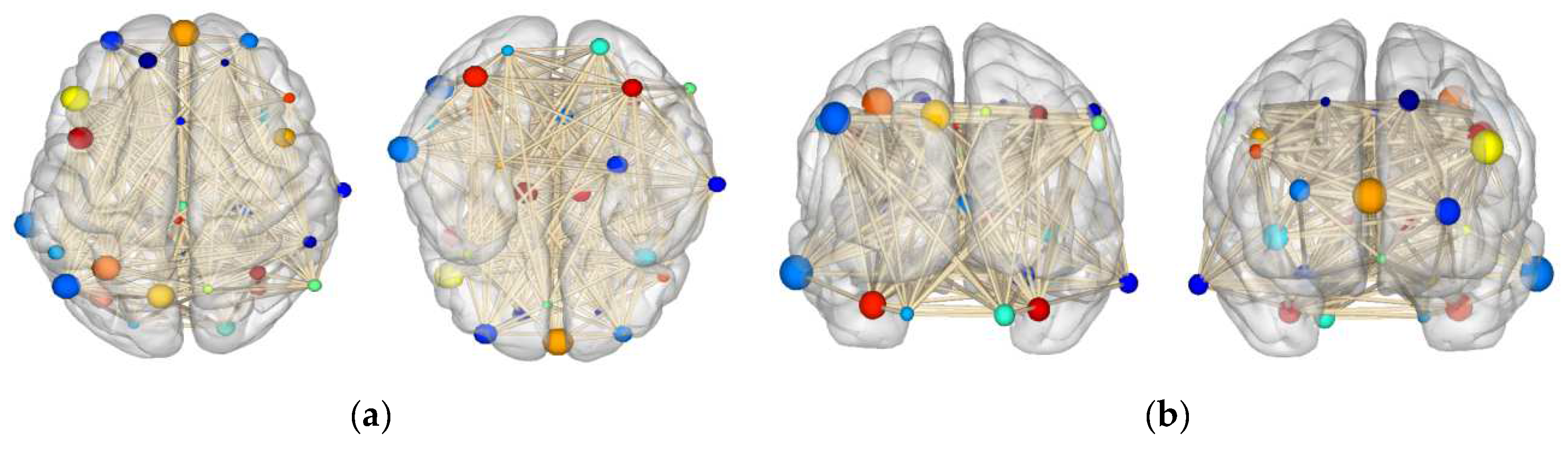
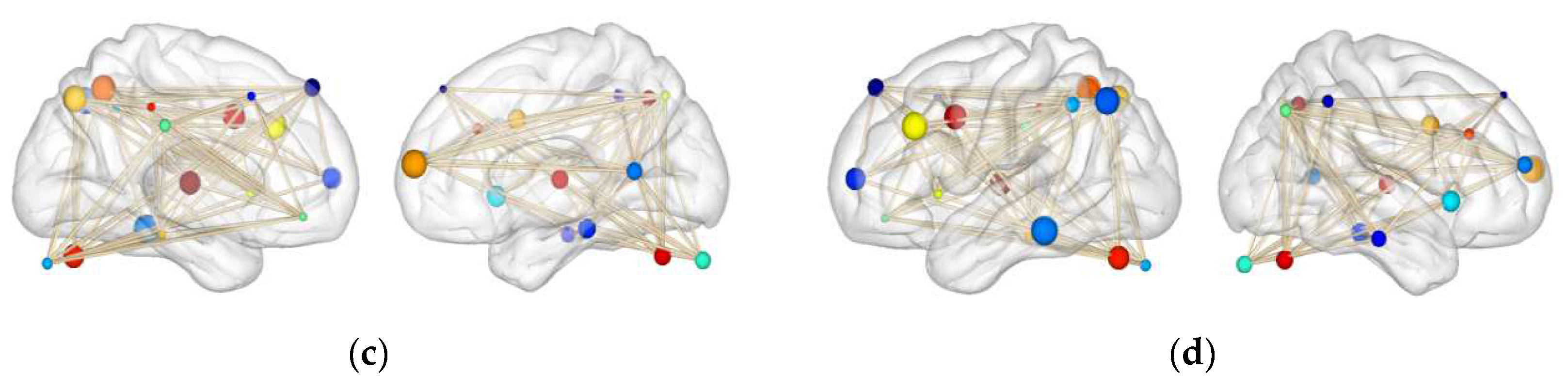
| Model | ISRUC | SHHS | ||||
|---|---|---|---|---|---|---|
| Accuracy | MF1 | Kappa | Accuracy | MF1 | Kappa | |
| MRCNN + AFR [25] | 0.606 | 0.552 | - | 0.689 | 0.557 | - |
| CNN + BiLSTM [10] | 0.788 | 0.779 | 0.730 | 0.719 | 0.588 | - |
| MLP + LSTM [12] | 0.779 | 0.713 | 0.758 | 0.802 | 0.779 | 0.792 |
| ResNet-50 [26] | 0.782 | - | 0.674 | 0.837 | - | 0.754 |
| ARNN + RNN [22] | 0.789 | 0.763 | 0.725 | 0.865 | 0.785 | 0.811 |
| STGCN [15] | 0.821 | 0.808 | 0.769 | - | - | - |
| proposed model | 0.825 | 0.814 | 0.775 | 0.873 | 0.801 | 0.827 |
| Model | Parameter | Training Duration |
|---|---|---|
| Supratak et al. [10] | 1.7 × 107 | 95.2 |
| Dong et al. [12] | 1.7 × 108 | 268 |
| Chambon et al. [11] | 2.0 × 105 | 9.98 |
| Phan et al. [22] | 1.2 × 105 | 367 |
| Chriskos et al. [23] | 4.1 × 105 | 75.8 |
| proposed model | 1.5 × 105 | 69.7 |
| Model Variable | Model Performance | ||
|---|---|---|---|
| Accuracy% | F1-Score% | Kappa% | |
| Baseline | 80.9 | 79.6 | 75.4 |
| Baseline + A | 81.5 | 80.3 | 76.2 |
| Baseline + B | 81.0 | 79.4 | 75.5 |
| Baseline + C | 81.4 | 80.3 | 76.1 |
| Baseline + A + B | 81.9 | 80.6 | 76.7 |
| Baseline + A + C | 82.4 | 81.1 | 77.3 |
| Baseline + B + C | 81.6 | 80.2 | 76.3 |
| Baseline + A + B + C(MGANet) | 82.5 | 81.4 | 77.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Guo, Y.; Shen, Y.; Tong, S.; Guo, H. Multi-Layer Graph Attention Network for Sleep Stage Classification Based on EEG. Sensors 2022, 22, 9272. https://doi.org/10.3390/s22239272
Wang Q, Guo Y, Shen Y, Tong S, Guo H. Multi-Layer Graph Attention Network for Sleep Stage Classification Based on EEG. Sensors. 2022; 22(23):9272. https://doi.org/10.3390/s22239272
Chicago/Turabian StyleWang, Qi, Yecai Guo, Yuhui Shen, Shuang Tong, and Hongcan Guo. 2022. "Multi-Layer Graph Attention Network for Sleep Stage Classification Based on EEG" Sensors 22, no. 23: 9272. https://doi.org/10.3390/s22239272
APA StyleWang, Q., Guo, Y., Shen, Y., Tong, S., & Guo, H. (2022). Multi-Layer Graph Attention Network for Sleep Stage Classification Based on EEG. Sensors, 22(23), 9272. https://doi.org/10.3390/s22239272





