Dual-Stage Deeply Supervised Attention-Based Convolutional Neural Networks for Mandibular Canal Segmentation in CBCT Scans
Abstract
:1. Introduction
2. Related Work
3. Materials and Methods
3.1. Study Design
3.2. Datasets
3.2.1. Our Dataset
3.2.2. Public Dataset
3.3. Data Pre-Processing
3.4. Overview of Dual-Stage Framework
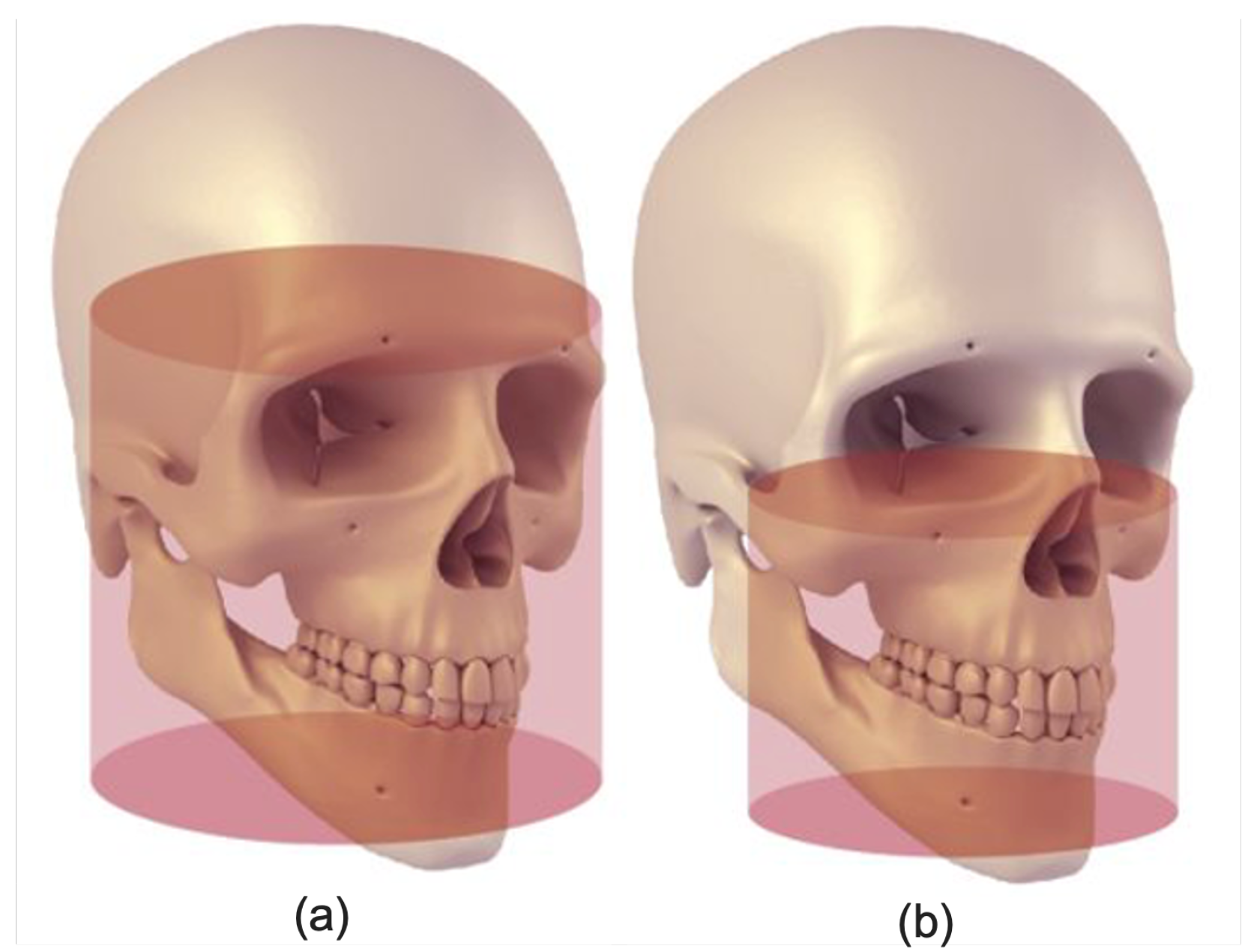
3.4.1. Jaw Localization
3.4.2. 3D Mandibular Canal Segmentation
3.5. Implementation Details and Training Strategy
3.6. Performance Measures
4. Results and Discussion
4.1. Benchmarking Results
4.2. Impact of Increasing the Amount of Data
4.3. Overall Performance Analysis
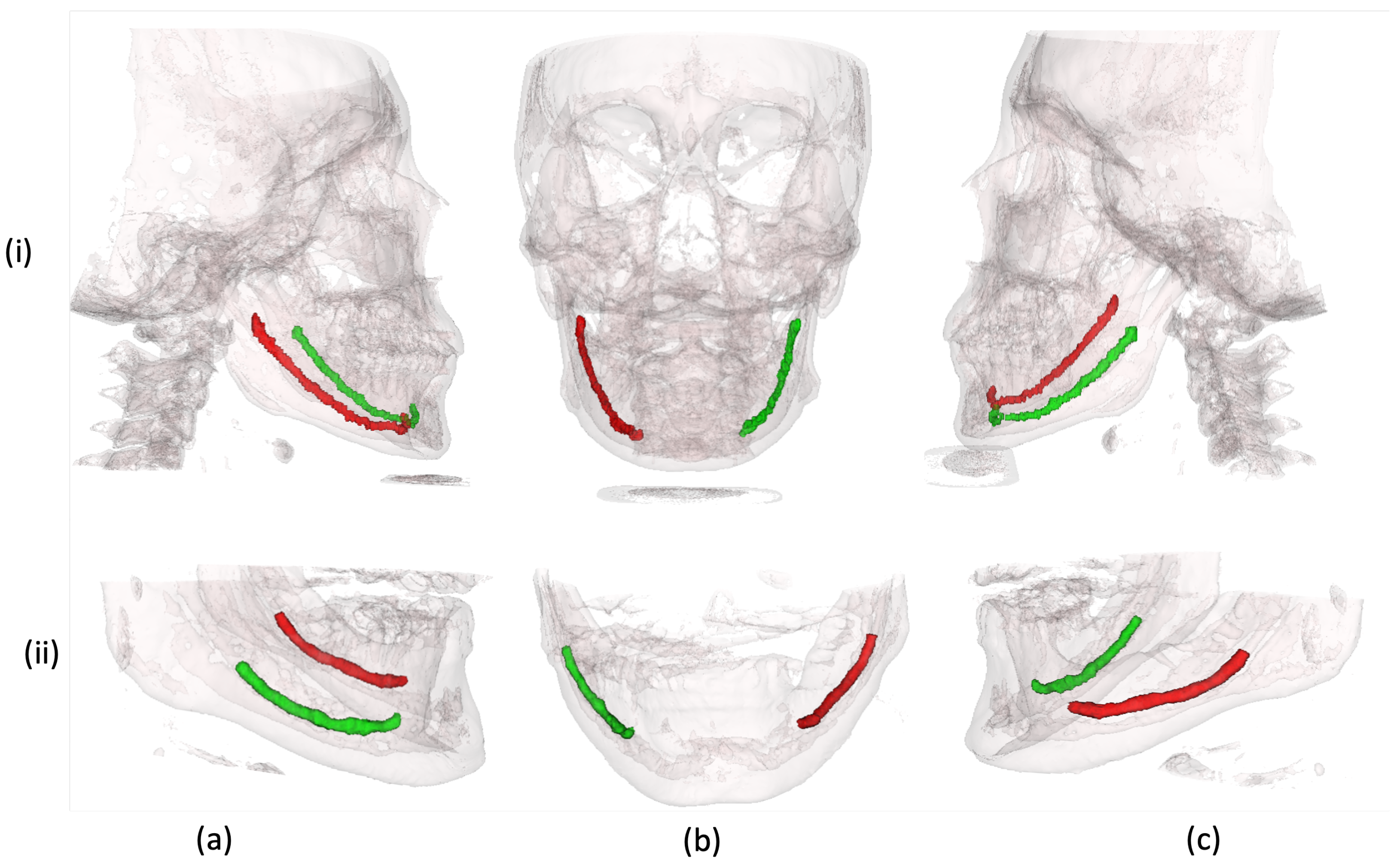
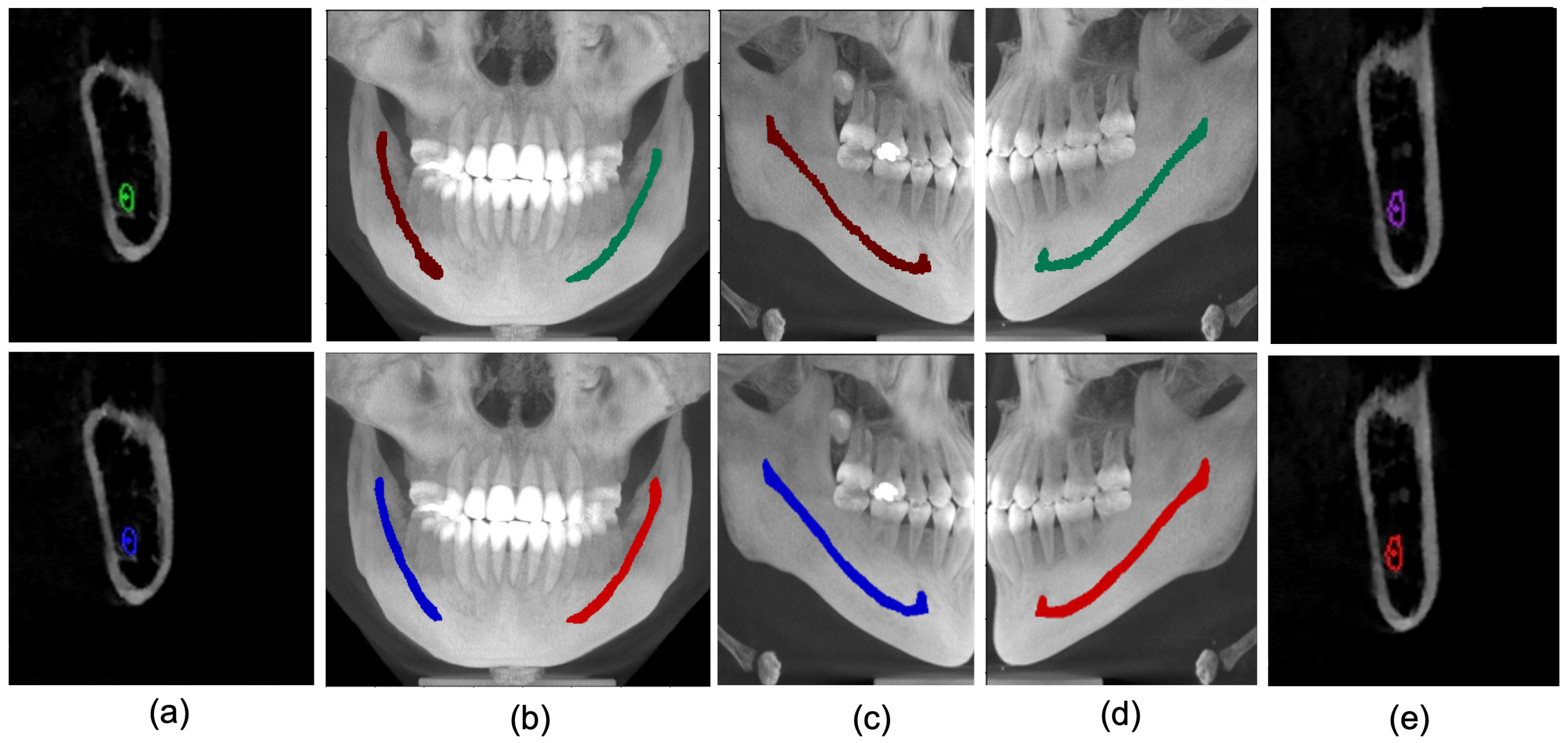
4.4. Qualitative Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Phillips, C.D.; Bubash, L.A. The facial nerve: Anatomy and common pathology. In Proceedings of the Seminars in Ultrasound, CT and MRI; Elsevier: Amsterdam, The Netherlands, 2002; Volume 23, pp. 202–217. [Google Scholar]
- Lee, E.G.; Ryan, F.S.; Shute, J.; Cunningham, S.J. The impact of altered sensation affecting the lower lip after orthognathic treatment. J. Oral Maxillofac. Surg. 2011, 69, e431–e445. [Google Scholar] [CrossRef] [PubMed]
- Juodzbalys, G.; Wang, H.L.; Sabalys, G. Injury of the inferior alveolar nerve during implant placement: A literature review. J. Oral Maxillofac. Res. 2011, 2, e1. [Google Scholar] [CrossRef] [PubMed]
- Westermark, A.; Zachow, S.; Eppley, B.L. Three-dimensional osteotomy planning in maxillofacial surgery including soft tissue prediction. J. Craniofacial Surg. 2005, 16, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.; Read-Fuller, A. Cone Beam Computed Tomography in Oral and Maxillofacial Surgery: An Evidence-Based Review. Dent. J. 2019, 7, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatcher, D.C. Operational principles for cone-beam computed tomography. J. Am. Dent. Assoc. 2010, 141, 3S–6S. [Google Scholar] [CrossRef]
- Angelopoulos, C.; Scarfe, W.C.; Farman, A.G. A comparison of maxillofacial CBCT and medical CT. Atlas Oral Maxillofac. Surg. Clin. N. Am. 2012, 20, 1–17. [Google Scholar] [CrossRef]
- Ghaeminia, H.; Meijer, G.; Soehardi, A.; Borstlap, W.; Mulder, J.; Vlijmen, O.; Bergé, S.; Maal, T. The use of cone beam CT for the removal of wisdom teeth changes the surgical approach compared with panoramic radiography: A pilot study. Int. J. Oral Maxillofac. Surg. 2011, 40, 834–839. [Google Scholar] [CrossRef]
- Chuang, Y.J.; Doherty, B.M.; Adluru, N.; Chung, M.K.; Vorperian, H.K. A novel registration-based semi-automatic mandible segmentation pipeline using computed tomography images to study mandibular development. J. Comput. Assist. Tomogr. 2018, 42, 306. [Google Scholar] [CrossRef]
- Wallner, J.; Hochegger, K.; Chen, X.; Mischak, I.; Reinbacher, K.; Pau, M.; Zrnc, T.; Schwenzer-Zimmerer, K.; Zemann, W.; Schmalstieg, D.; et al. Clinical evaluation of semi-automatic open-source algorithmic software segmentation of the mandibular bone: Practical feasibility and assessment of a new course of action. PLoS ONE 2018, 13, e0196378. [Google Scholar] [CrossRef]
- Issa, J.; Olszewski, R.; Dyszkiewicz-Konwińska, M. The Effectiveness of Semi-Automated and Fully Automatic Segmentation for Inferior Alveolar Canal Localization on CBCT Scans: A Systematic Review. Int. J. Environ. Res. Public Health 2022, 19, 560. [Google Scholar] [CrossRef]
- Kim, G.; Lee, J.; Lee, H.; Seo, J.; Koo, Y.M.; Shin, Y.G.; Kim, B. Automatic extraction of inferior alveolar nerve canal using feature-enhancing panoramic volume rendering. IEEE Trans. Biomed. Eng. 2010, 58, 253–264. [Google Scholar]
- Abdolali, F.; Zoroofi, R.A.; Abdolali, M.; Yokota, F.; Otake, Y.; Sato, Y. Automatic segmentation of mandibular canal in cone beam CT images using conditional statistical shape model and fast marching. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 581–593. [Google Scholar] [CrossRef]
- Abdolali, F.; Zoroofi, R.A.; Biniaz, A. Fully automated detection of the mandibular canal in cone beam CT images using Lie group based statistical shape models. In Proceedings of the 25th IEEE National and 3rd International Iranian Conference on Biomedical Engineering (ICBME), Qom, Iran, 29–30 November 2018; pp. 1–6. [Google Scholar]
- Wei, X.; Wang, Y. Inferior alveolar canal segmentation based on cone-beam computed tomography. Med. Phys. 2021, 48, 7074–7088. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.S.; Kim, Y.S.; Kim, M.; Usman, M.; Byon, S.S.; Kim, S.H.; Lee, B.I.; Lee, B.D. Evaluation of the feasibility of explainable computer-aided detection of cardiomegaly on chest radiographs using deep learning. Sci. Rep. 2021, 11, 16885. [Google Scholar] [CrossRef] [PubMed]
- Usman, M.; Lee, B.D.; Byon, S.S.; Kim, S.H.; Lee, B.i.; Shin, Y.G. Volumetric lung nodule segmentation using adaptive roi with multi-view residual learning. Sci. Rep. 2020, 10, 12839. [Google Scholar] [CrossRef] [PubMed]
- Kwak, G.H.; Kwak, E.J.; Song, J.M.; Park, H.R.; Jung, Y.H.; Cho, B.H.; Hui, P.; Hwang, J.J. Automatic mandibular canal detection using a deep convolutional neural network. Sci. Rep. 2020, 10, 5711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaskari, J.; Sahlsten, J.; Järnstedt, J.; Mehtonen, H.; Karhu, K.; Sundqvist, O.; Hietanen, A.; Varjonen, V.; Mattila, V.; Kaski, K. Deep learning method for mandibular canal segmentation in dental cone beam computed tomography volumes. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Faradhilla, Y.; Arifin, A.Z.; Suciati, N.; Astuti, E.R.; Indraswari, R.; Widiasri, M. Residual Fully Convolutional Network for Mandibular Canal Segmentation. Int. J. Intell. Eng. Syst. 2021, 14, 208–219. [Google Scholar]
- Widiasri, M.; Arifin, A.Z.; Suciati, N.; Fatichah, C.; Astuti, E.R.; Indraswari, R.; Putra, R.H.; Za’in, C. Dental-YOLO: Alveolar Bone and Mandibular Canal Detection on Cone Beam Computed Tomography Images for Dental Implant Planning. IEEE Access 2022, 10, 101483–101494. [Google Scholar] [CrossRef]
- Albahli, S.; Nida, N.; Irtaza, A.; Yousaf, M.H.; Mahmood, M.T. Melanoma lesion detection and segmentation using YOLOv4-DarkNet and active contour. IEEE Access 2020, 8, 198403–198414. [Google Scholar] [CrossRef]
- Dhar, M.K.; Yu, Z. Automatic tracing of mandibular canal pathways using deep learning. arXiv 2021, arXiv:2111.15111. [Google Scholar]
- Verhelst, P.J.; Smolders, A.; Beznik, T.; Meewis, J.; Vandemeulebroucke, A.; Shaheen, E.; Van Gerven, A.; Willems, H.; Politis, C.; Jacobs, R. Layered deep learning for automatic mandibular segmentation in cone-beam computed tomography. J. Dent. 2021, 114, 103786. [Google Scholar] [CrossRef] [PubMed]
- Lahoud, P.; Diels, S.; Niclaes, L.; Van Aelst, S.; Willems, H.; Van Gerven, A.; Quirynen, M.; Jacobs, R. Development and validation of a novel artificial intelligence driven tool for accurate mandibular canal segmentation on CBCT. J. Dent. 2022, 116, 103891. [Google Scholar] [CrossRef] [PubMed]
- Cipriano, M.; Allegretti, S.; Bolelli, F.; Di Bartolomeo, M.; Pollastri, F.; Pellacani, A.; Minafra, P.; Anesi, A.; Grana, C. Deep segmentation of the mandibular canal: A new 3D annotated dataset of CBCT volumes. IEEE Access 2022, 10, 11500–11510. [Google Scholar] [CrossRef]
- Cipriano, M.; Allegretti, S.; Bolelli, F.; Pollastri, F.; Grana, C. Improving segmentation of the inferior alveolar nerve through deep label propagation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 21137–21146. [Google Scholar]
- Du, G.; Tian, X.; Song, Y. Mandibular Canal Segmentation From CBCT Image Using 3D Convolutional Neural Network With scSE Attention. IEEE Access 2022, 10, 111272–111283. [Google Scholar] [CrossRef]
- Technology. Digital Radiographic Images in Dental Practice. Available online: https://www.lauc.net/en/technology/ (accessed on 14 December 2022).
- Hochreiter, S. The vanishing gradient problem during learning recurrent neural nets and problem solutions. Int. J. Uncertain. Fuzziness Knowl.-Based Syst. 1998, 6, 107–116. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Zhang, J.; Ruan, C.; Zhang, Y. Multi-Stage Attention-Unet for Wireless Capsule Endoscopy Image Bleeding Area Segmentation. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 818–825. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, Q. Road Extraction by Deep Residual U-Net. IEEE Geosci. Remote. Sens. Lett. 2017, 15, 749–753. [Google Scholar] [CrossRef] [Green Version]
- Soille, P. Erosion and Dilation. In Morphological Image Analysis: Principles and Applications; Springer: Berlin/Heidelberg, Germany, 2004; pp. 63–137. [Google Scholar] [CrossRef]
- Chollet, F. Keras. 2015. Available online: https://github.com/fchollet/keras (accessed on 14 December 2022).
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. 2015. Available online: tensorflow.org (accessed on 14 December 2022).




| Author, Study and Year of Publication | Technique | Type of Dataset and FOV | No. of CBCT Scans | Contributions | Limitations | |
|---|---|---|---|---|---|---|
| Training + Validation | Test | |||||
| Kwak et al. [19], 2020 | Thresholding-based teeth segmentation + 3D UNets | Private, Full View | 82 | 20 | Employed 2D and 3D Deep Learning models and demonstrated the superior performance of 3D UNets | Limited performance in terms of Mean mIoU |
| Jaskari et al. [20], 2020 | 3D Fully Convolutional Neural Networks (FCNNs) | Private, Medium View | 509 | 128 | The study utilized a large number of CBCT scans to train 3D FCNNs and achieved an improved performance. | Overall achieved performance for left and right canal was limited in term of Dice score |
| Faradhilla et al. [21], 2021 | Residual FCNNs + Dual auxilary Loss functions | Private, 2D view | NA | NA | The study exploited Residual Fully Convolutional Network with dual auxiliary loss functions to segment the mandibular canal in parasagittal 2D images and reported promising results in terms of dice score | Requires manual input from dentists to generate the 2D parasagittal views from CBCT. Study provides no information about the CBCT scans used for the experimentation |
| Verhelst et al. [25], 2021 | 3D UNet trained in two phases | Private, Medium View | 160 | 30 | Trained 3D UNet in two phases, i.e., before and after the deployment, to achieve promising performance. | Requires an extensive effort to train the model and inputs from experts are needed to improve its performance of the model. |
| Widiasri et al. [22], 2022 | YOLOv4 | Private, 2D view | NA | NA | The study utilized YOLOv4 for mandibular canal detection in 2D coronal images and achieved significantly higher detection performance. | The study used 2D coronal images which need manual input to generate from CBCT scans. The technique just provides the bounding box around the canal region, which lacks the exact boundary information which can be obtained from segmentation. |
| Lahoud et al. [26], 2022 | Two 3D UNets, one for coarse segmentation and other for finetuning on patches | Private, Mixed FOVs | 196 | 39 | Adjusted to the variability in Mandibular Canal shape and width by using voxel-wise probability approach for segmentation | The scheme requires an extensive effort to train the models and evaluate performed on a limited private dataset does not prove the generalization |
| Cipriano et al. [27], 2022 | Jaskari et al. [20], 2020 | Public, Medium view | 76 with Dense annotation | 15 with Dense Annotations | The first publicly released annotated dataset and source code, validated their dataset on three different existing techniques | Utilized the existing segmentation methods, with no contribution in terms of technique novelty. |
| Cipriano et al. [28], 2022 | 3D CNN + Deep label propagation technique | Public, Medium view | 76 with Dense annotation+256 with Sparse Annotations | 15 with Dense Annotations | Combined 3D segmentation model trained on the 3D annotated data and label propagation model to improve the mandibular canal segmentation performance | The study utilized the scans with a medium Field Of View (FOV) which is 3D sub-volume from CBCT scans, however, no mechanism for localization of medium FOV is provided. |
| Our Dataset | Public Dataset [27] | |
|---|---|---|
| Total Number of CBCT scans | 1010 | 347 |
| Densely annotated scans | 1010 | 91 |
| Sparsely Annotated scans | - | 256 |
| Minimum Resolution | 512 × 512 × 460 | 148 × 265 × 312 |
| Maximum Resolution | 670 × 670 × 640 | 178 × 423 × 463 |
| Pixel Spacing | 0.3 mm–0.39 mm | 0.3 mm |
| Field of View | Large | Medium |
| Performance Parameters | Without Multi-Scale | Without Resiudual Connections | With Residual Connections and Multi-Scale Inputs |
|---|---|---|---|
| mIoU | 0.779 | 0.785 | 0.795 |
| Precision | 0.683 | 0.679 | 0.69 |
| Recall | 0.81 | 0.824 | 0.83 |
| Dice Score | 0.72 | 0.72 | 0.751 |
| F1 Score | 0.741 | 0.745 | 0.759 |
| Number of Scans | 100 | 200 | 300 | 400 | ||||
|---|---|---|---|---|---|---|---|---|
| Canal Side | Left | Right | Left | Right | Left | Right | Left | Right |
| mIoU | 0.755 | 0.771 | 0.78 | 0.789 | 0.79 | 0.8 | 0.798 | 0.806 |
| Precision | 0.639 | 0.667 | 0.657 | 0.69 | 0.679 | 0.718 | 0.686 | 0.72 |
| Recall | 0.818 | 0.795 | 0.832 | 0.8 | 0.847 | 0.817 | 0.854 | 0.819 |
| Dice Score | 0.721 | 0.731 | 0.734 | 0.746 | 0.749 | 0.753 | 0.752 | 0.759 |
| F1 Score | 0.718 | 0.725 | 0.734 | 0.741 | 0.754 | 0.764 | 0.761 | 0.766 |
| Study | Field of View | Training Scans | Testing Scans | Solution Type | mIoU | Precision | Recall | Dice Score | F1 Score |
|---|---|---|---|---|---|---|---|---|---|
| Jaskari et al. [20], 2020 | Medium | 457 | 128 | Automated | - | - | - | 0.575 | - |
| Kwak et al. [19], 2020 (3D) | Medium | 61 | - | Automated | 0.577 | - | - | - | - |
| Dhar et al. [24], 2021 | Medium | 157 | 30 | Automated | 0.7 | 0.63 | 0.51 | - | 0.56 |
| Verhelst et al. [25], 2021 | Medium | 196 | 39 | Semi-Automated | 0.946 | 0.952 | 0.993 | 0.972 | - |
| Lahoud et al. [26], 2022 | Medium + Large | 166 | 39 | Automated | 0.636 | 0.782 | 0.792 | 0.774 | - |
| 100 | 500 | Automated | 0.763 | 0.653 | 0.807 | 0.726 | 0.721 | ||
| Our method | Large | 200 | 500 | Automated | 0.785 | 0.67 | 0.816 | 0.74 | 0.737 |
| 300 | 500 | Automated | 0.795 | 0.69 | 0.832 | 0.751 | 0.759 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Usman, M.; Rehman, A.; Saleem, A.M.; Jawaid, R.; Byon, S.-S.; Kim, S.-H.; Lee, B.-D.; Heo, M.-S.; Shin, Y.-G. Dual-Stage Deeply Supervised Attention-Based Convolutional Neural Networks for Mandibular Canal Segmentation in CBCT Scans. Sensors 2022, 22, 9877. https://doi.org/10.3390/s22249877
Usman M, Rehman A, Saleem AM, Jawaid R, Byon S-S, Kim S-H, Lee B-D, Heo M-S, Shin Y-G. Dual-Stage Deeply Supervised Attention-Based Convolutional Neural Networks for Mandibular Canal Segmentation in CBCT Scans. Sensors. 2022; 22(24):9877. https://doi.org/10.3390/s22249877
Chicago/Turabian StyleUsman, Muhammad, Azka Rehman, Amal Muhammad Saleem, Rabeea Jawaid, Shi-Sub Byon, Sung-Hyun Kim, Byoung-Dai Lee, Min-Suk Heo, and Yeong-Gil Shin. 2022. "Dual-Stage Deeply Supervised Attention-Based Convolutional Neural Networks for Mandibular Canal Segmentation in CBCT Scans" Sensors 22, no. 24: 9877. https://doi.org/10.3390/s22249877
APA StyleUsman, M., Rehman, A., Saleem, A. M., Jawaid, R., Byon, S.-S., Kim, S.-H., Lee, B.-D., Heo, M.-S., & Shin, Y.-G. (2022). Dual-Stage Deeply Supervised Attention-Based Convolutional Neural Networks for Mandibular Canal Segmentation in CBCT Scans. Sensors, 22(24), 9877. https://doi.org/10.3390/s22249877






