Concept of Fluorescent Transport Activity Biosensor for the Characterization of the Arabidopsis NPF1.3 Activity of Nitrate
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Constructs
2.2. Yeast Cultures
2.3. Fluorimetry
2.4. Functional Expression of NPF1.3 in Xenopus Oocytes
2.5. Electrophysiological Measurements in Xenopus Oocytes
2.6. Statistical Analyses
3. Results
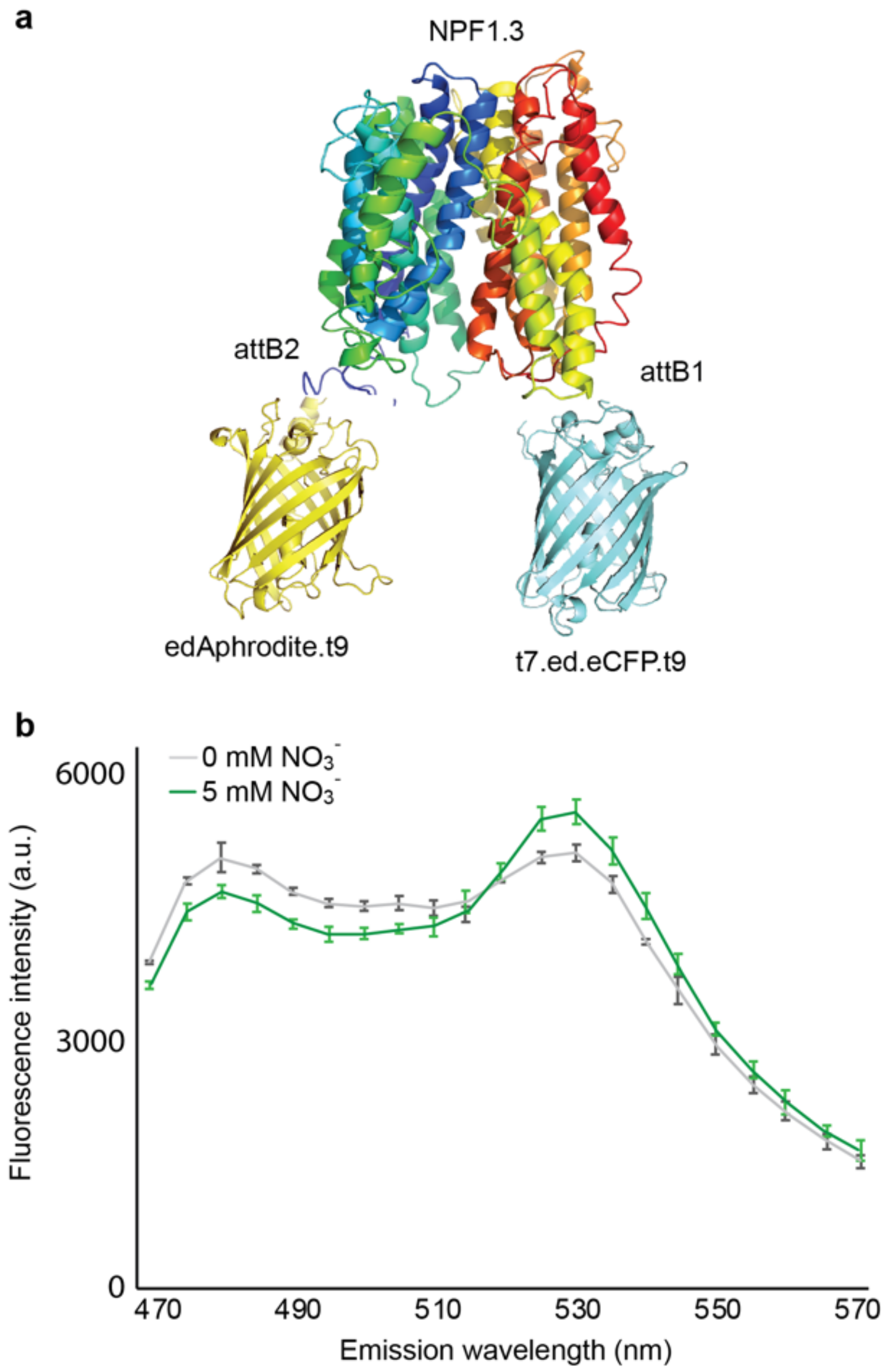
3.1. A Screen of Engineering of Members of NPF into Transport Activity Sensor
3.2. Selectivity of NiTrac-NPF1.3 Biosensor
3.3. A Single Cell Measurement of NiTrac-NPF1.3 Biosensor
3.4. NPF1.3 Functions as Nitrate Transporter
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- McAllister, C.H.; Beatty, P.H.; Good, A.G. Engineering nitrogen use efficient crop plants: The current status. Plant Biotechnol. J. 2012, 10, 1011–1025. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Stewart, G.R. Nitrate Assimilation and Translocation by Higher-Plants—Comparative Physiology and Ecological Consequences. Physiol. Plant. 1985, 64, 133–140. [Google Scholar] [CrossRef]
- Song, J.; Ding, X.D.; Feng, G.; Zhang, F.S. Nutritional and osmotic roles of nitrate in a euhalophyte and a xerophyte in saline conditions. New Phytol. 2006, 171, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G. Local and long-range signaling pathways regulating plant responses to nitrate. Annu. Rev. Plant Biol. 2002, 53, 203–224. [Google Scholar] [CrossRef]
- Sakakibara, H.; Takei, K.; Hirose, N. Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci. 2006, 11, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Gutierrez, R.A. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr. Opin. Plant Biol. 2008, 11, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Okumoto, S. Quantitative imaging using genetically encoded sensors for small molecules in plants. Plant J. 2012, 70, 108–117. [Google Scholar] [CrossRef] [Green Version]
- De Michele, R.; Ast, C.; Loque, D.; Ho, C.H.; Andrade, S.L.; Lanquar, V.; Grossmann, G.; Gehne, S.; Kumke, M.U.; Frommer, W.B. Fluorescent sensors reporting the activity of ammonium transceptors in live cells. eLife 2013, 2, e00800. [Google Scholar] [CrossRef]
- Deuschle, K.; Chaudhuri, B.; Okumoto, S.; Lager, I.; Lalonde, S.; Frommer, W.B. Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell 2006, 18, 2314–2325. [Google Scholar] [CrossRef] [Green Version]
- Chaudhuri, B.; Hörmann, F.; Frommer, W.B. Dynamic imaging of glucose flux impedance using FRET sensors in wild-type Arabidopsis plants. J. Exp. Bot. 2011, 62, 2411–2417. [Google Scholar] [CrossRef] [Green Version]
- Chaudhuri, B.; Hörmann, F.; Lalonde, S.; Brady, S.M.; Orlando, D.A.; Benfey, P.; Frommer, W.B. Protonophore- and pH-insensitive glucose and sucrose accumulation detected by FRET nanosensors in Arabidopsis root tips. Plant J. 2008, 56, 948–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okumoto, S.; Takanaga, H.; Frommer, W.B. Quantitative imaging for discovery and assembly of the metabo-regulome. New Phytol. 2008, 180, 271–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doki, S.; Kato, H.E.; Solcan, N.; Iwaki, M.; Koyama, M.; Hattori, M.; Iwase, N.; Tsukazaki, T.; Sugita, Y.; Kandori, H.; et al. Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT. Proc. Natl. Acad. Sci. USA 2013, 110, 11343–11348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, C.H.; Frommer, W.B. Fluorescent sensors for activity and regulation of the nitrate transceptor CHL1/NRT1.1 and oligopeptide transporters. eLife 2014, 3, e01917. [Google Scholar] [CrossRef] [PubMed]
- Ast, C.; Foret, J.; Oltrogge, L.M.; De Michele, R.; Kleist, T.J.; Ho, C.H.; Frommer, W.B. Ratiometric Matryoshka biosensors from a nested cassette of green- and orange-emitting fluorescent proteins. Nat. Commun. 2017, 8, 431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddiqi, M.Y.; Glass, A.D.M.; Ruth, T.J.; Rufty, T.W., Jr. Studies of the uptake of nitrate in barley. I. Kinetics of 13NO3− influx. Plant Physiol. 1990, 93, 1426–1432. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.Y.; Glass, A.; Shaff, J.E.; Kochian, L.V. Ammonium Uptake by Rice Roots (III. Electrophysiology). Plant Physiol. 1994, 104, 899–906. [Google Scholar] [CrossRef] [Green Version]
- von Wirén, N.; Gazzarrini, S.; Frommer, W.B. Regulation of mineral nitrogen uptake in plants. In Plant Nutrition—For Sustainable Food Production and Environment; Kluwer Academic Publishers: Amsterdam, The Netherlands, 1997; pp. 41–49. [Google Scholar]
- Leran, S.; Varala, K.; Boyer, J.C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B.; et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2013, 19, 5–9. [Google Scholar] [CrossRef]
- Kotur, Z.; Mackenzie, N.; Ramesh, S.; Tyerman, S.D.; Kaiser, B.N.; Glass, A.D. Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytol. 2012, 194, 724–731. [Google Scholar] [CrossRef]
- Lin, S.H.; Kuo, H.F.; Canivenc, G.; Lin, C.S.; Lepetit, M.; Hsu, P.K.; Tillard, P.; Lin, H.L.; Wang, Y.Y.; Tsai, C.B.; et al. Mutation of the Arabidopsis NRT1.5 Nitrate Transporter Causes Defective Root-to-Shoot Nitrate Transport. Plant Cell 2008, 20, 2514–2528. [Google Scholar] [CrossRef] [Green Version]
- Li, J.Y.; Fu, Y.L.; Pike, S.M.; Bao, J.; Tian, W.; Zhang, Y.; Chen, C.Z.; Zhang, Y.; Li, H.M.; Huang, J.; et al. The Arabidopsis Nitrate Transporter NRT1.8 Functions in Nitrate Removal from the Xylem Sap and Mediates Cadmium Tolerance. Plant Cell 2010, 22, 1633–1646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, S.C.; Lin, C.S.; Hsu, P.K.; Lin, S.H.; Tsay, Y.F. The Arabidopsis Nitrate Transporter NRT1.7, Expressed in Phloem, Is Responsible for Source-to-Sink Remobilization of Nitrate. Plant Cell 2009, 21, 2750–2761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.Y.; Tsay, Y.F. Arabidopsis Nitrate Transporter NRT1.9 Is Important in Phloem Nitrate Transport. Plant Cell 2011, 23, 1945–1957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.Y.; Lin, S.H.; Cheng, L.H.; Wu, J.J.; Lin, Y.C.; Tsay, Y.F. Potential transceptor AtNRT1.13 modulates shoot architecture and flowering time in a nitrate-dependent manner. Plant Cell 2021, 33, 1492–1505. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.K.; Tsay, Y.F. Two Phloem Nitrate Transporters, NRT1.11 and NRT1.12, Are Important for Redistributing Xylem-Borne Nitrate to Enhance Plant Growth. Plant Physiol. 2013, 163, 844–856. [Google Scholar] [CrossRef] [Green Version]
- Chiu, C.C.; Lin, C.S.; Hsia, A.P.; Su, R.C.; Lin, H.L.; Tsay, Y.F. Mutation of a nitrate transporter, AtNRT1: 4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 2004, 45, 1139–1148. [Google Scholar] [CrossRef]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef]
- Kanno, Y.; Hanada, A.; Chiba, Y.; Ichikawa, T.; Nakazawa, M.; Matsui, M.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. USA 2012, 109, 9653–9658. [Google Scholar] [CrossRef] [Green Version]
- Boursiac, Y.; Leran, S.; Corratge-Faillie, C.; Gojon, A.; Krouk, G.; Lacombe, B. ABA transport and transporters. Trends Plant Sci. 2013, 18, 325–333. [Google Scholar] [CrossRef]
- Jones, A.M.; Grossmann, G.; Danielson, J.A.; Sosso, D.; Chen, L.Q.; Ho, C.H.; Frommer, W.B. In vivo biochemistry: Applications for small molecule biosensors in plant biology. Curr. Opin. Plant Biol. 2013, 16, 389–395. [Google Scholar] [CrossRef] [Green Version]
- Loqué, D.; Mora, S.I.; Andrade, S.L.; Pantoja, O.; Frommer, W.B. Pore mutations in ammonium transporter AMT1 with increased electrogenic ammonium transport activity. J. Biol. Chem. 2009, 284, 24988–24995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, C.-H.; Frommer, W.B. Design and Functional Analysis of Fluorescent Nitrate and Peptide Transporter Activity Sensors in Yeast Cultures. Bio-Protocol 2016, 6, e1728. [Google Scholar] [CrossRef]
- Gietz, D.; Jean, A.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef] [PubMed]
- Bermejo, C.; Haerizadeh, F.; Takanaga, H.; Chermak, D.; Frommer, W.B. Dynamic analysis of cytosolic glucose and ATP levels in yeast using optical sensors. Biochem. J. 2010, 432, 399–406. [Google Scholar] [CrossRef] [Green Version]
- Bermejo, C.; Haerizadeh, F.; Takanaga, H.; Chermak, D.; Frommer, W.B. Optical sensors for measuring dynamic changes of cytosolic metabolite levels in yeast. Nat. Protoc. 2011, 6, 1806–1817. [Google Scholar] [CrossRef]
- Bermejo, C.; Ewald, J.C.; Lanquar, V.; Jones, A.M.; Frommer, W.B. In vivo biochemistry: Quantifying ion and metabolite levels in individual cells or cultures of yeast. Biochem. J. 2011, 438, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Pehl, U.; Leisgen, C.; Gampe, K.; Guenther, E. Automated higher-throughput compound screening on ion channel targets based on the Xenopus laevis oocyte expression system. Assay Drug Dev. Technol. 2004, 2, 515–524. [Google Scholar] [CrossRef]
- Lemaire, K.; Van de Velde, S.; Van Dijck, P.; Thevelein, J.M. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 2004, 16, 293–299. [Google Scholar] [CrossRef] [Green Version]
- Huang, N.C.; Liu, K.H.; Lo, H.J.; Tsay, Y.F. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 1999, 11, 1381–1392. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, M.A.; Springer, G.; Segawa, K.; Zipfel, W.R.; Piston, D.W. Optimization of pairings and detection conditions for measurement of FRET between cyan and yellow fluorescent proteins. Microsc. Microanal. 2006, 12, 238–254. [Google Scholar] [CrossRef]
- He, Y.N.; Peng, J.S.; Cai, Y.; Liu, D.F.; Guan, Y.; Yi, H.Y.; Gong, J.M. Tonoplast-localized nitrate uptake transporters involved in vacuolar nitrate efflux and reallocation in Arabidopsis. Sci. Rep. 2017, 7, 6417. [Google Scholar] [CrossRef] [PubMed]
- Almagro, A.; Lin, S.H.; Tsay, Y.F. Characterization of the Arabidopsis Nitrate Transporter NRT1.6 Reveals a Role of Nitrate in Early Embryo Development. Plant Cell 2008, 20, 3289–3299. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Byrt, C.; Qiu, J.; Baumann, U.; Hrmova, M.; Evrard, A.; Johnson, A.A.T.; Birnbaum, K.D.; Mayo, G.M.; Jha, D.; et al. Identification of a Stelar-Localized Transport Protein That Facilitates Root-to-Shoot Transfer of Chloride in Arabidopsis. Plant Physiol. 2016, 170, 1014–1029. [Google Scholar] [CrossRef]
- Li, B.; Qiu, J.; Jayakannan, M.; Xu, B.; Li, Y.; Mayo, G.M.; Tester, M.; Gilliham, M.; Roy, S.J. AtNPF2.5 Modulates Chloride (Cl-) Efflux from Roots of Arabidopsis thaliana. Front. Plant Sci. 2017, 7, 2013. [Google Scholar] [CrossRef] [Green Version]
- Branco-Price, C.; Kawaguchi, R.; Ferreira, R.B.; Bailey-Serres, J. Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann. Bot. 2005, 96, 647–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammes, U.Z.; Meier, S.; Dietrich, D.; Ward, J.M.; Rentsch, D. Functional properties of the Arabidopsis peptide transporters AtPTR1 and AtPTR5. J. Biol. Chem. 2010, 285, 39710–39717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komarova, N.Y.; Thor, K.; Gubler, A.; Meier, S.; Dietrich, D.; Weichert, A.; Suter Grotemeyer, M.; Tegeder, M.; Rentsch, D. AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiol. 2008, 148, 856–869. [Google Scholar] [CrossRef]
- Chiang, C.S.; Stacey, G.; Tsay, Y.F. Mechanisms and functional properties of two peptide transporters, AtPTR2 and fPTR2. J. Biol. Chem. 2004, 279, 30150–30157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Yu, M.; Du, X.Q.; Wang, Z.F.; Wu, W.H.; Quintero, F.J.; Jin, X.H.; Li, H.D.; Wang, Y. NRT1.5/NPF7.3 Functions as a Proton-Coupled H+/K+ Antiporter for K+ Loading into the Xylem in Arabidopsis. Plant Cell 2017, 29, 2016–2026. [Google Scholar] [CrossRef] [Green Version]
- Tal, I.; Zhang, Y.; Jorgensen, M.E.; Pisanty, O.; Barbosa, I.C.R.; Zourelidou, M.; Regnault, T.; Crocoll, C.; Olsen, C.E.; Weinstain, R.; et al. The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 2016, 7, 11486. [Google Scholar] [CrossRef]
- Nour-Eldin, H.H.; Andersen, T.G.; Burow, M.; Madsen, S.R.; Jorgensen, M.E.; Olsen, C.E.; Dreyer, I.; Hedrich, R.; Geiger, D.; Halkier, B.A. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 2012, 488, 531–534. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-N.; Ho, C.-H. Concept of Fluorescent Transport Activity Biosensor for the Characterization of the Arabidopsis NPF1.3 Activity of Nitrate. Sensors 2022, 22, 1198. https://doi.org/10.3390/s22031198
Chen Y-N, Ho C-H. Concept of Fluorescent Transport Activity Biosensor for the Characterization of the Arabidopsis NPF1.3 Activity of Nitrate. Sensors. 2022; 22(3):1198. https://doi.org/10.3390/s22031198
Chicago/Turabian StyleChen, Yen-Ning, and Cheng-Hsun Ho. 2022. "Concept of Fluorescent Transport Activity Biosensor for the Characterization of the Arabidopsis NPF1.3 Activity of Nitrate" Sensors 22, no. 3: 1198. https://doi.org/10.3390/s22031198





