1. Introduction
Among the various cancerous diseases that affect humanity all across the world, breast cancer is the second leading cause of cancer deaths after lung cancer [
1]. This type of cancer, if not detected in the early stage, might lead to death, and accounts for a large number of fatalities among women [
2]. Early detection and diagnosis can have high chances of successful treatment of this disease and decrease the physical and mental pain endured by patients.
There are many biomedical imaging techniques for the early detection of Breast Cancer such as, Digital Mammography, Ultrasound, and MRI based diagnoses [
3]. However, most of the methods have serious radiation effects. Moreover, some of these tests do not even confirm the malignancy of the cancerous tissues. In such scenarios, Breast Biopsy is performed to confirm the same. A biopsy is a histopathological assessment of the microscopic structure of the tissues. The biopsy can be used to differentiate between normal tissue, benign, and malignant tissues [
4]. The only disadvantage of using histopathological images is zooming and focusing on the required part, which is highly time-consuming and demands well-experienced pathologists. This is where Computer-Aided Diagnosis (CAD) plays its role, providing a highly accurate classification of those tissue images, thus providing a second opinion to the doctors [
5].
With the approach of machine learning in the field of Breast Cancer Histopathology image analysis, the detection in the early stages of cancer has proved to be an excellent research area. Deep Learning, one of the most recent machine learning methods, has outperformed the old and conventional methods in many image analysis tasks [
6]. The most common Deep Learning algorithms include Convolutional Neural Networks(CNNs). The application of convolutional neural networks for pattern recognition and feature extraction in medical imaging have proved to be quite successful [
7]. Moreover, studies have shown that fine-tuning of pre-trained models using CNN as the base can achieve comparatively higher performance in a wide variety of medical imaging tasks. The knowledge from a pre-trained source can be used to improve the learning of the actual model, thereby increasing the performance, which is way better than traditional CNN techniques. CNNs are the structural representation of a stream of feature extraction stages across its layers. The knowledge obtained by training with image samples is primarily fed as the weights of the layers. However, in Transfer Learning models, one common strategy is to freeze the weights from the input layer up to a particular layer to use those feature extraction layers from the pre-trained weights [
8]. Moreover, in Transfer Learning techniques, the models require less computational power and data as compared to Deep CNN models altogether, reducing training time. The only problem that arises is that transfer learning models suffer from low generalization capability and might lead to overfitting [
9].
In this study, a transfer learning based approach is being implemented where a pre-trained CNN model will be used along with some fine-tuned layers to obtain the target task. This methodology has been implemented in breast cancer imaging since 2016, following the development of several pre-trained CNN models, including VGGNet, Inception, and ResNet, to solve visual classification tasks. The ImageNet image database would be used for the pre-trained weights [
10]. The proposed work summarizes existing methods and identifies their performances on breast cancer detection.
1.1. Problem Statement
Presently, breast cancer diagnoses are performed majorly by various biomedical imaging techniques. However, these techniques involve the use of radiation, which might cause adverse health effects in the future. Thus only breast tissue biopsy can be treated as an effective method for diagnoses of the same. Moreover distinguishing between the malignant and normal tissues is also a hectic task when the number of patients is unexpectedly high. So, this paper proposes an android application for quick and reliable diagnoses of breast cancer wherein the user can easily upload an image and get results aided by deep learning.
1.2. Contributions of the Proposed Paper
The main contribution of ABCanDroid is:
A Cloud integrated Android app for the detection of breast cancer from a Breast Histopathology image;
The proposed work used a transfer learning based model, trained with 15,616 images and tested on 3904 images for the prediction of breast cancer;
The model has accuracy and precision of 99.58% while tested on the 3904 images (Whole Slide Images);
The proposed framework is of low cost and requires very minimal human intervention and as it is cloud integrated, so less performance load on the edge devices.
1.3. Article Structure
The rest of the paper is organized as follows:
Section 2 as Related work.
Section 3 is the Proposed Methodology,
Section 4 is Experimental Results. Finally,
Section 5 presents Conclusion and Future Works.
2. Related Works
Various Machine and Deep learning methods have been implemented in the field of health care systems (see
Table 1). Most of the works implemented on Breast cancer detection are related to binary or multi-class classification using machine learning or deep learning. These works include performance report parameters such as accuracy, precision, recall, and F1 score. This section portrays some of the works related to Breast Cancer detection and classification.
In a work by Rakhlin et al. [
11], a deep learning based system was proposed using pre-trained models such as VGG16, InceptionV3, and ResNet50 for the classification of images of breast tissue. The pre-trained models were used for feature extraction. This technique achieved an accuracy of 87.2% across image classification. In another work by Kwok et al. [
12], four Deep Convolutional Neural Network(DCNN) architectures have been used for classification. They tried to increase the accuracy by several data augmentation techniques. Vang et al. [
13] proposed an ensemble-learning based architecture for multi-class classification problems in breast cancer. Their classifier involved using logistic regression and Gradient boosting machine. These models failed to achieve the high accuracy, which is essential for any sort of medical imaging classification.
In a paper by Nawaz et al. [
14], they implemented a fine-tuned AlexNet for the classification of breast cancer. They achieved an accuracy of 75.73% when they used a patch-wise dataset. Xu et al. [
15] proposed a CNN based model for segmenting breast ultrasound images into four major tissues where the accuracy of their model reached over 80%. Fang et al. [
16] proposed a novel method based on ultrasound images for breast cancer classification. Their segmentation algorithm received an accuracy of 88%. Again, in these above works the accuracy is well below 90%. Authors also have not provided any measure of incorporating the models in edge devices, which can be made readily available to users for easy diagnoses.
In another work by Reza et al. [
17], they considered imbalanced data and worked upon them using Convolutional Neural Networks with different layers. They also used image data augmentation to create altered versions of the same image. They achieved an accuracy of about 85% in their work. Similar to this work, Ali et al. [
18] proposed a neural architecture with a color constancy technique and achieved an accuracy of 93.5%. Their work also involved histogram equalization but its accuracy was not that good. Wang et al. [
19] also used a similar technique of using CNNs with an added batch normalization layer. They employed five different types of models with several layers and came up with an efficient one that achieved an accuracy of 89%. The idea of using transfer learning in a similar work was adopted by Singh et al. [
20], where they worked on imbalanced data from WSI dataset and used VGG-19 with different classifiers like logistic regression, random forest, and other dense layers.
In the work by Alzubaidi et al. [
21], they presented a thorough study of how pre-training Transfer Learning models with different datasets can affect the performance of the model. In their study, the authors proved that pre-training models with a similar kind of data can significantly improve the model’s performance. In another work by Azizi et al. [
22], they presented a novel Multi-Instance Contrastive Learning (MICLe) method, which makes use of pretrained ImageNet model followed by self-supervised learning on multiple unlabeled medical images of pathology of the same patient. Later, they fine-tuned the model with labelled medical images. This method greatly improved the accuracy of their model by 6–7% against existing models.
In the work by Ayana et al. [
23], they focused on transfer learning methods applied on ultrasound breast image classification and detection using pre-trained CNN models. Moreover, their review on some of the most commonly used transfer learning techniques presents the potential of future research in this field. Khamparia et al. [
24] in their work proposed a hybrid transfer learning model, mainly a fusion of MVGG and ImageNet, and achieved an accuracy of 94.3%. They implemented their model on the WSI dataset. They also employed image segmentation and 3D mammography in their work, which helped to acquire better results.
Table 1.
Comparative study of related works for breast cancer detection.
Table 1.
Comparative study of related works for breast cancer detection.
| Author | Works | Salient Features | Transfer Learning | Application on Edge Devices |
|---|
| Samala et al. [25] | Multi-Stage Transfer Learning used with Deep Neural Nets | Applicable for limited data and gain in performance | ✓ | × |
| Choudhary et al. [26] | Transfer Learning based on Structural Filter Pruning Approach | Applicable for point-of-care devices | ✓ | × |
| Deniz et al. [27] | Transfer Learning with deep feature extraction from pretrained models | Applicable for outperforming traditional ML models | ✓ | × |
| Ayana et al. [23] | Transfer Learning technique on Ultrasound Images | Applicable for better image processing | ✓ | × |
| Khamparia et al. [24] | Implemented Hybrid Transfer Learning | Applicable for increasing efficiency of model | ✓ | × |
| Zhang et al. [28] | Implemented combination of Transfer Learning and Recurrent Neural Net | Applicable for better result outcome | ✓ | × |
| Alzubaidi et al. [29] | Implemented DCNN based Double Transfer Learning model on histopathological images | Applicable for limited labelled data for classification and high precision | ✓ | × |
| Gatuha et al. [30] | Cloud based Android app for breast cancer detection using Naive Bayes classification | Applicable for ease of use | × | ✓ |
| ABCanDroid | Android based Transfer Learning Implementation on Histopathological Images | Applicable for ease of use and highly precise accuracy | ✓ | ✓ |
In another work by Choudhary et al. [
26], they performed thorough experiments using three popular pre-trained CNNs such as VGG19, ResNet34, and ResNet50. With the use of the VGG19 pruned model, they obtained an accuracy of 91.25%, outperforming initial methods on the same dataset. Sheikh et al. [
31] made in their work a thorough comparison among six different classifier levels along with deep learning based algorithms. They inferred that some of their algorithms enhanced the performance of Breast Cancer classification to a large extent. Deniz et al. [
27] used Fine Tuned AlexNet and VGG16 transfer learning models, achieving an accuracy of 91.3%. Both of the model features were extracted and concatenated for better results. In the above mentioned works, the authors have succeeded in achieving a moderately high accuracy, but the authors did not provide any method for making the model available for common use. These drawbacks have been addressed in this work. The work proposed in this paper is a cloud integrated transfer learning based android app. This facilities the app, being light-weight solving problems related to computation. Additionally, integrating the model with android app helps facilitate its availability to masses. These previously discussed papers have implemented their work on the same WSI Dataset and have gained good results.
Lastly, Alzubaidi et al. [
29] provided a novel approach to resolve the issue of lack of data in medical imaging. The authors pre-trained DCNN model on a large number of unlabeled histopathological images of breast cancer. After fine tuning, the model was trained with small labeled dataset of breast cancer. This process enabled the authors to achieve an accuracy of 97.51%. The authors also applied novel double transfer learning, achieving an accuracy of 97.7%.
Gatuha et al. [
30] provided a cloud based android application for breast cancer detection. The authors provided a data mining technique based on Naïve Bayes probabilistic classifier for breast cancer detection from images. They obtained an accuracy of 96.4%. In this paper, a Transfer Learning based model has been used, which produces significantly better results as compared to traditional machine learning algorithms.
Critical Analysis
The comparison of the proposed work with all other transfer learning and DNN based models is presented in
Table 1. The research works presented above have shown quite a good performance in the detection of breast cancer when measured in terms of accuracy, but it is also necessary to implement it in a manner that would be available to patients in general. Out of all the presented works, only a few have employed the use of transfer learning. Considering the works done in terms of implementing a transfer learning based Android App, there is a requirement for a framework that gives better accuracy as well as facilitate the users with an Android Application, as shown in
Figure 1.
In such a scenario, there is a need to improve the current framework benchmarks: (1) to build an association between users and the proposed framework, (2) to incorporate the best possible procedure to achieve greater accuracy, and (3) to provide an ideal experience to both patients and doctors. This proposed framework tries to improve the current scenario of existing frameworks with this research work.
3. Proposed Methodology
In this section, the description of the proposed approach used to achieve the objectives of the work is discussed, covering system architecture and datasets used to CNN based classification. The methodology adopted involves the use of transfer learning on the input histopathological images.
3.1. System Architecture
The proposed system takes an input of a Breast Histopathological Image to identify Breast Cancer. First of all, this system converts images taken from the user, from the current colour channel to the Red-Green-Blue (RGB) color channel. Furthermore, the system will consider only images that are similar to Breast Histopathological Images. First, the image is quantified through a Structural Similarity Index (SSIM) measure to check its structural similarity with a Histopathological image. Then, only the image is used for detection or classification.
Figure 2 depicts the sequence of steps involving input from dataset, splitting data into testing and training and finally, after implementation, on four DNN Models and predicting the output labels. The ResNet model is pretty effective in the extraction of features and classifying images based on those features. The whole model was pushed to a serverless cloud service [
32] named Heroku and then the results were fetched using an API from the Android application backend. The Android App was developed using Flutter Mobile Framework paired with the API fetching service. This was done so as to prevent the loss of accuracy when TfLite was used with Flutter.
3.2. Data Cleaning and Input Preprocessing
Before the model is trained, there is a requirement of cleaning the dataset and converting it to the proper form to be fed into deep neural networks for classification. Image processing is a necessary step to achieve significant and accurate classification. The database includes images of various sizes ranging from 512 × 512 pixels to 1024 × 1024 pixels. Therefore, before passing the images for feature extraction, they have been resized to a size of 224 × 224 × 3 pixels to be ready as input to the system. The preprocessing needed for applying transfer learning on breast cancer histopathological images also involves reducing class imbalance. Moreover, the models were also trained on original sized images without any cropping or resizing to ensure there is no loss in quality or features from the image. A comparison chart of the accuracy across 5 folds of BreakHis Dataset is visualized in
Figure 3.
Then, the images were labeled correspondingly using a label list that had 0 as Normal and 1 as Affected classes. Thus, the problem decreases to binary classification. The image list was then converted into a NumPy array and the dimensions were reduced within a range of 0 to 1 by dividing with 255. Then, the whole dataset was divided into Training and Testing images and labels with a ratio of 80:20, respectively.
3.3. Transfer Learning Approach
In medical imaging problems there is usually a scarcity of data. To overcome this problem, transfer learning has come into play and has helped to deal with small data and achieve better performance. In medical cases, especially in breast cancer imaging, different types of images are used for classification. Some of the popular types include Magnetic Resonance Imaging (MRI), Computed Tomography (CT), and Ultrasound (US). However, in this approach, the use of Histopathological images have been employed to detect and classify breast cancer images.
3.3.1. Feature Extraction
In general, there are two conventional approaches for transfer learning, namely feature extraction and fine-tuning. In the feature extraction approach, a well trained CNN model on a large dataset such as ImageNet is used to extract the features of the target domain, for example, in breast cancer imaging. The convolutional layers of the pretrained model are used as a frozen feature extractor to match with a new classification task. These features are then sent to a classifier, which is trained throughout the training process of the entire network. The feature map of the learned features obtained from a sample image when passed through the neural network architecture is visualized in
Figure 4. These feature maps are from the base functional ResNet101 Layer along with the First Convolutional Layer of the neural network. The feature maps give an insight into the edges and features of the input images as it passes through various layers.
3.3.2. Pretrained Model Dataset
The most common pretrained models used for transfer learning include ResNet, DensNet, Inception, and so on. Out of these, the InceptionV3 model is quite common. In this work, a comparative study of the different pre-trained models was employed to find out the best of the lot. In breast cancer imaging based on transfer learning, the ImageNet dataset is commonly used.
The ImageNet dataset is a large image database designed for image recognition tasks. It generally consists of 14 million images that have been annotated to identify pictured objects. This dataset is capable of classifying more than 20,000 categories with a particular category consisting of various images.
3.4. Transfer Learning CNN Models
A CNN in breast cancer image analysis is basically a feed-forward neural network. The main advantage of using CNN is its accuracy in image recognition tasks. However, it requires high computational power and huge training data. A CNN usually consists of a base input layer along with pooling and convolutional layers. Finally there is a fully connected layer, which is the output layer providing the classification results. Some of the most commonly used CNN models for transfer learning with breast cancer images are the following:
3.4.1. VGG16
VGG16 was the first CNN introduced by Visual Geometry Group (VGG). VGG16 is a type of convolution neural network that consists of 13 convolution neural networks and 3 fully connected layers. This was further followed by VGG19. These architectures were based on ImageNet dataset. The main feature of VGG16 is transfer learning based CNNs used as fixed feature extractor. It is a pre-trained CNN architecture trained on a large dataset, where the last fully connected layer of this pre-trained network is removed. The remaining CNN acts as a fixed feature extractor for the new dataset.
3.4.2. DenseNet
DenseNet is a more recent architecture that is used in image classification problems. It shows exceptional performances in terms of classification accuracy, despite having a fewer number of parameters. Advantages of DenseNet include parameter efficiency, in which every layer adds only a limited number of parameters. On the other side, it is more helpful because it has a higher capacity with multi-layer feature concatenation. DenseNets obtain significant improvements over the state-of-the-art on most of them, whilst requiring less memory and computation to achieve high performance.
3.4.3. Xception
Xception is a convolutional neural network. It was first introduced by Google researchers. They used their idea on Xception from the Inception model because depthwise separable convolutional was better than Inception. When data is inserted to be classified, then at first it enters into the entry flow. Then, it goes through the middle flow and finally enters from the exit flow. It is a useful architecture that works on Depthwise Separable Convolution and thus makes shortcuts between convolutional blocks. Xception consists of 36 convolutional layers, and these layers take part to form the feature extraction base of networks.
3.4.4. ResNet
This paper discusses three different CNN techniques—VGG16, DenseNet, and Xception, respectively. In this study, the ResNet based transfer learning technique has been implemented, which will contain ImageNet data weights downloaded from the web. Instead of building a deep learning model from scratch, a more practical approach was adopted, constructing a model using already proven models. The main advantage of ResNet model is the presence of a large number of layers. Transfer learning enables us to retrain the final layer of an existing model, resulting in a significant decrease in training time. One of the most famous models that can be used for transfer learning is ResNet.
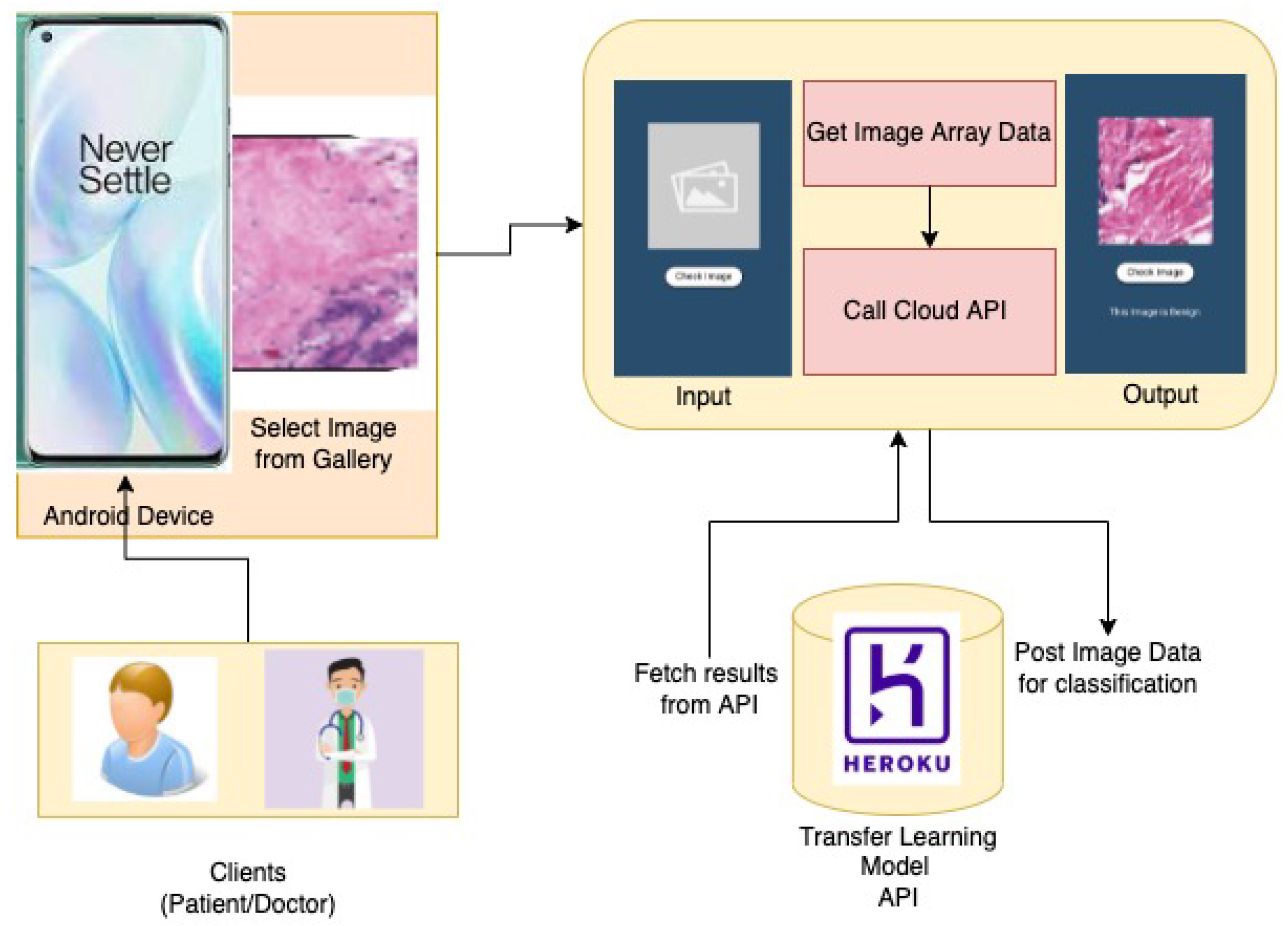
3.5. The Prototype Application
Following the proposed approach, a cloud integrated mobile application has been developed based on the Android operating system for the detection and classification of Breast Cancer Histopathological Images.The App Architecture Design is shown in
Figure 5 The app allows users to browse and upload an image and feed it to the application. The application in turn will evaluate the image, using the model proposed and provide a classified label probability. The proposed application can be used by patients having biopsy or histopathological images obtained from clinics. In addition, doctors can utilize this application for easy diagnoses of breast cancer and also to save their time for fast and efficient diagnoses.
6. Conclusions
The main goal of ABCanDroid was to create an improved breast cancer classification system that would be affordable and accessible to various healthcare systems. Considering the limitations of primitive machine learning models, the proposed study purposely used the pretrained models to extract some fine-tuned features before training on histopathological images. Several extensive experiments were performed on the four pre-trained transfer learning models, namely VGG16, DenseNet121, Xception, and finally Resnet101.
The comparative study, as shown in
Table 9, indicates the superiority of transfer learning. Transfer Learning has aided the development of breast cancer diagnoses by overcoming the challenge of obtaining a large training dataset. Apart from these various preprocessing techniques, such as colour conversion, augmentation has also played a significant role in improving the performance of the model.
ABCanDroid helps users to distinguish between malignant and normal tissues by uploading a single histopathological image at a time. The proposed model has delivered an accuracy of 99.58%. In this paper, the main focus is on breast cancer detection using an android app.