FRDD-Net: Automated Carotid Plaque Ultrasound Images Segmentation Using Feature Remapping and Dense Decoding
Abstract
:1. Introduction
- (1)
- To mitigate challenge 1, a novel feature remapping module is proposed. FRMs embedded in encoding and decoding blocks can reweight input features to facilitate their rationality.
- (2)
- To mitigate challenge 2, a novel dense decoding mechanism is proposed. Such decoding architecture can exploit hierarchical features along with their fusions to promote segmentation performance.
- (3)
- To mitigate challenge 3, a novel compound loss function is constructed. The loss function can improve FRDD-Net’s reliability when handling intractable cases.
2. Related Works
2.1. Traditional Methods for the Carotid Ultrasound Image Segmentation
2.2. Deep Neural Networks for the Segmentation of Carotid Plaque Ultrasound Image
3. Materials and Methods
3.1. Data Preprocessing
3.2. Overall Architecture
3.3. Feature Remapping Module
3.4. Dense Decoding Mechanism
3.5. Compound Loss Function
4. Results and Discussions
4.1. Dataset and Implementation Details
4.2. Qualitative and Quantitative Analysis of Carotid Plaque Segmentation
4.3. Cross-Dataset Studies
4.4. Ablation Studies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. World Health Statistics 2019: Monitoring Health for the SDGs, Sustainable Development Goals; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Feigin, V.L.; Forouzanfar, M.H.; Krishnamurthi, R.; Mensah, G.A.; Connor, M.; Bennett, D.A.; Moran, A.E.; Sacco, R.L.; Anderson, L.; Truelsen, T.; et al. Global and regional burden of stroke during 1990–2010: Findings from the Global Burden of Disease Study 2010. Lancet 2014, 383, 245–255. [Google Scholar] [CrossRef]
- Lo, E.H.; Dalkara, T.; Moskowitz, M.A. Mechanisms, challenges and opportunities in stroke. Nat. Rev. Neurosci. 2003, 4, 399–414. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Sharma, N.; Giannopoulos, A.A.; Saba, L.; Nicolaides, A.; Suri, J.S. Hybrid deep learning segmentation models for atherosclerotic plaque in internal carotid artery B-mode ultrasound. Comput. Biol. Med. 2021, 136, 104721. [Google Scholar] [CrossRef] [PubMed]
- Buda, N.; Segura-Grau, E.; Cylwik, J.; Wełnicki, M. Lung ultrasound in the diagnosis of COVID-19 infection-A case series and review of the literature. Adv. Med Sci. 2020, 65, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Wagner, W.R. Non-invasive and non-destructive characterization of tissue engineered constructs using ultrasound imaging technologies: A review. Ann. Biomed. Eng. 2016, 44, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Loizou, C.P.; Petroudi, S.; Pattichis, C.S.; Pantziaris, M.; Kasparis, T.; Nicolaides, A. Segmentation of atherosclerotic carotid plaque in ultrasound video. In Proceedings of the 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Diego, CA, USA, 28 August–1 September 2012; pp. 53–56. [Google Scholar]
- Destrempes, F.; Soulez, G.; Giroux, M.F.; Meunier, J.; Cloutier, G. Segmentation of plaques in sequences of ultrasonic B-mode images of carotid arteries based on motion estimation and Nakagami distributions. In Proceedings of the 2009 IEEE International Ultrasonics Symposium, Rome, Italy, 20–23 September 2009; pp. 2480–2483. [Google Scholar]
- Akkus, Z.; De Jong, N.; Van Der Steen, A.F.; Bosch, J.G.; Van Den Oord, S.C.; Schinkel, A.F.; Carvalho, D.D.; Niessen, W.J.; Klein, S. Fully automated carotid plaque segmentation in combined b-mode and contrast enhanced ultrasound. In Proceedings of the 2014 IEEE International Ultrasonics Symposium, Chicago, IL, USA, 3–6 September 2014; pp. 911–914. [Google Scholar]
- Loizou, C.P.; Petroudi, S.; Pantziaris, M.; Nicolaides, A.N.; Pattichis, C.S. An integrated system for the segmentation of atherosclerotic carotid plaque ultrasound video. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2014, 61, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Li, Y.; Xue, Y.; Huntress, L.; Beckerman, W.; Rahimi, S.A.; Ady, J.W.; Roshan, U.W. Two-stage and dual-decoder convolutional U-Net ensembles for reliable vessel and plaque segmentation in carotid ultrasound images. In Proceedings of the 2020 19th IEEE International Conference on Machine Learning and Applications (ICMLA), Miami, FL, USA, 14–17 December 2020; pp. 1376–1381. [Google Scholar]
- Azzopardi, C.; Camilleri, K.P.; Hicks, Y.A. Bimodal automated carotid ultrasound segmentation using geometrically constrained deep neural networks. IEEE J. Biomed. Health Inform. 2020, 24, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Fenster, A.; Xia, Y.; Spence, J.D.; Ding, M. Deep learning-based carotid media-adventitia and lumen-intima boundary segmentation from three-dimensional ultrasound images. Med. Phys. 2019, 46, 3180–3193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, M.A. Neural Networks and Deep Learning; Determination Press: San Francisco, CA, USA, 2015; Volume 25. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Menchón-Lara, R.M.; Sancho-Gómez, J.L. Fully automatic segmentation of ultrasound common carotid artery images based on machine learning. Neurocomputing 2015, 151, 161–167. [Google Scholar] [CrossRef]
- Shin, J.; Tajbakhsh, N.; Hurst, R.T.; Kendall, C.B.; Liang, J. Automating carotid intima-media thickness video interpretation with convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2526–2535. [Google Scholar]
- Vila, M.d.M.; Remeseiro, B.; Grau, M.; Elosua, R.; Igual, L. Last Advances on Automatic Carotid Artery Analysis in Ultrasound Images: Towards Deep Learning. In Handbook of Artificial Intelligence in Healthcare; Springer: Berlin/Heidelberg, Germany, 2022; pp. 215–247. [Google Scholar]
- Sumathi, K.; Mahesh, V.; Ramakrishnan, S. Analysis of intima media thickness in ultrasound carotid artery images using level set segmentation without re-initialization. In Proceedings of the 2014 International Conference on Informatics, Electronics & Vision (ICIEV), Dhaka, Bangladesh, 23–24 May 2014; pp. 1–4. [Google Scholar]
- Carvalho, D.D.; Akkus, Z.; Van Den Oord, S.C.; Schinkel, A.F.; Van Der Steen, A.F.; Niessen, W.J.; Bosch, J.G.; Klein, S. Lumen segmentation and motion estimation in B-mode and contrast-enhanced ultrasound images of the carotid artery in patients with atherosclerotic plaque. IEEE Trans. Med. Imaging 2014, 34, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Guo, F.; Azarpazhooh, M.R.; Spence, J.D.; Ukwatta, E.; Ding, M.; Fenster, A. A voxel-based fully convolution network and continuous max-flow for carotid vessel-wall-volume segmentation from 3D ultrasound images. IEEE Trans. Med. Imaging 2020, 39, 2844–2855. [Google Scholar] [CrossRef] [PubMed]
- Azzopardi, C.; Hicks, Y.A.; Camilleri, K.P. Automatic carotid ultrasound segmentation using deep convolutional neural networks and phase congruency maps. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, Australia, 18–21 April 2017; pp. 624–628. [Google Scholar]
- Perez, E.; Strub, F.; De Vries, H.; Dumoulin, V.; Courville, A. Film: Visual reasoning with a general conditioning layer. In Proceedings of the AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018; Volume 32. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Li, X.; Sun, X.; Meng, Y.; Liang, J.; Wu, F.; Li, J. Dice loss for data-imbalanced NLP tasks. arXiv 2019, arXiv:1911.02855. [Google Scholar]
- Abraham, N.; Khan, N.M. A novel focal tversky loss function with improved attention u-net for lesion segmentation. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 683–687. [Google Scholar]
- Salehi, S.S.M.; Erdogmus, D.; Gholipour, A. Tversky loss function for image segmentation using 3D fully convolutional deep networks. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2017; pp. 379–387. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019, 32, 8026–8037. [Google Scholar]
- Loshchilov, I.; Hutter, F. Sgdr: Stochastic gradient descent with warm restarts. arXiv 2016, arXiv:1608.03983. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]







| Method | Baseline | DSC (%) | IoU (%) |
|---|---|---|---|
| PSPNet | efficientnet-b0 | 75.76 | 65.72 |
| DeepLabV3 | efficientnet-b0 | 82.18 | 75.68 |
| DeepLabV3+ | efficientnet-b0 | 81.36 | 74.37 |
| U-net | efficientnet-b0 | 82.39 | 76.05 |
| U-net++ | efficientnet-b0 | 82.19 | 75.71 |
| FRDD-Net | efficientnet-b0 | 83.20 | 77.41 |
| FRDD-Net | FR-encoder | 83.65 | 78.18 |
| Method | Baseline | DSC (%) | IoU (%) |
|---|---|---|---|
| PSPNet | efficientnet-b0 | 68.56 | 55.47 |
| DeepLabV3 | efficientnet-b0 | 71.69 | 59.41 |
| DeepLabV3+ | efficientnet-b0 | 71.15 | 59.58 |
| U-net | efficientnet-b0 | 77.73 | 66.80 |
| U-net++ | efficientnet-b0 | 80.54 | 68.24 |
| FRDD-Net | FR-encoder | 82.61 | 70.69 |
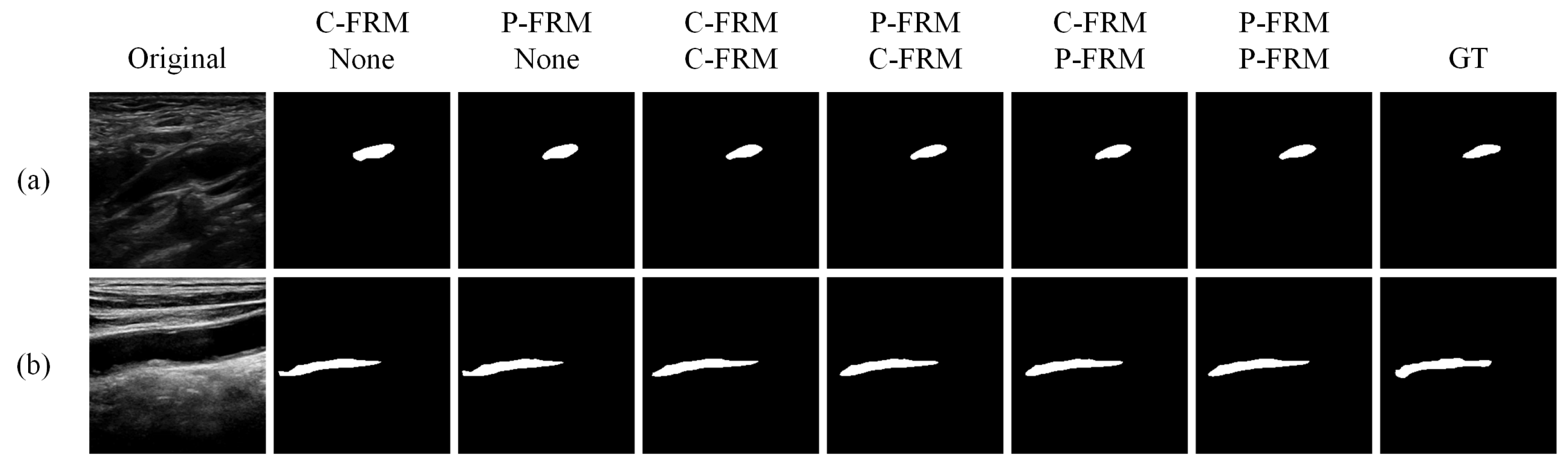
| Encoder | Decoder | DSC (%) | IoU (%) |
|---|---|---|---|
| C-FRM | None | 82.23 | 75.80 |
| P-FRM | None | 82.46 | 76.18 |
| C-FRM | C-FRM | 83.26 | 77.51 |
| P-FRM | C-FRM | 83.59 | 78.06 |
| C-FRM | P-FRM | 83.54 | 78.00 |
| P-FRM | P-FRM | 83.65 | 78.18 |
| Loss Function | DSC (%) | IoU (%) |
|---|---|---|
| 82.29 | 75.88 | |
| 83.65 | 78.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zou, L.; Xiong, L.; Yu, F.; Jiang, H.; Fan, C.; Cheng, M.; Li, Q. FRDD-Net: Automated Carotid Plaque Ultrasound Images Segmentation Using Feature Remapping and Dense Decoding. Sensors 2022, 22, 887. https://doi.org/10.3390/s22030887
Li Y, Zou L, Xiong L, Yu F, Jiang H, Fan C, Cheng M, Li Q. FRDD-Net: Automated Carotid Plaque Ultrasound Images Segmentation Using Feature Remapping and Dense Decoding. Sensors. 2022; 22(3):887. https://doi.org/10.3390/s22030887
Chicago/Turabian StyleLi, Yanhan, Lian Zou, Li Xiong, Fen Yu, Hao Jiang, Cien Fan, Mofan Cheng, and Qi Li. 2022. "FRDD-Net: Automated Carotid Plaque Ultrasound Images Segmentation Using Feature Remapping and Dense Decoding" Sensors 22, no. 3: 887. https://doi.org/10.3390/s22030887
APA StyleLi, Y., Zou, L., Xiong, L., Yu, F., Jiang, H., Fan, C., Cheng, M., & Li, Q. (2022). FRDD-Net: Automated Carotid Plaque Ultrasound Images Segmentation Using Feature Remapping and Dense Decoding. Sensors, 22(3), 887. https://doi.org/10.3390/s22030887






