1. Introduction
Machine learning, especially Deep Learning (DL) approaches, has been of interest in academia and industry. This has resulted in numerous changes in the approach to automatic detection or classification processes. However, the reliability of such studies has not always been high and differs depending on the methods used.
Since recently, it has been proved that Artificial Intelligence (AI) and machine learning has numerous applications in all engineering fields. Among them are the areas of electrical engineering [
1], civil engineering [
2], and petroleum engineering [
3]. In addition, classification using DL methods [
4] have several practical applications in various areas of medicine, such as the diagnosis of diseases based on physiological parameters [
5], the classification of cardiac arrhythmias based on ECG signals [
6,
7], and the recognition of human activity [
8]. Various ECG classification schemes based on DL were used to detect heart diseases [
9,
10,
11,
12], for example, using Long Short-Term Memory networks [
13] and one-dimensional Convolution Neural Networks [
14,
15,
16]. In addition, DL methods have been used to classify pathological conditions of the heart, such as arrhythmia, atrial fibrillation, ventricular fibrillation, and others.
Cardiovascular disease is a general term for a series of cardiovascular abnormalities that are the world’s leading cause of death [
17]. Each of them is identified and interpreted using an electrocardiogram (ECG). The ECG is an important non-invasive diagnostic method for the interpretation and identification of various types of heart disease.
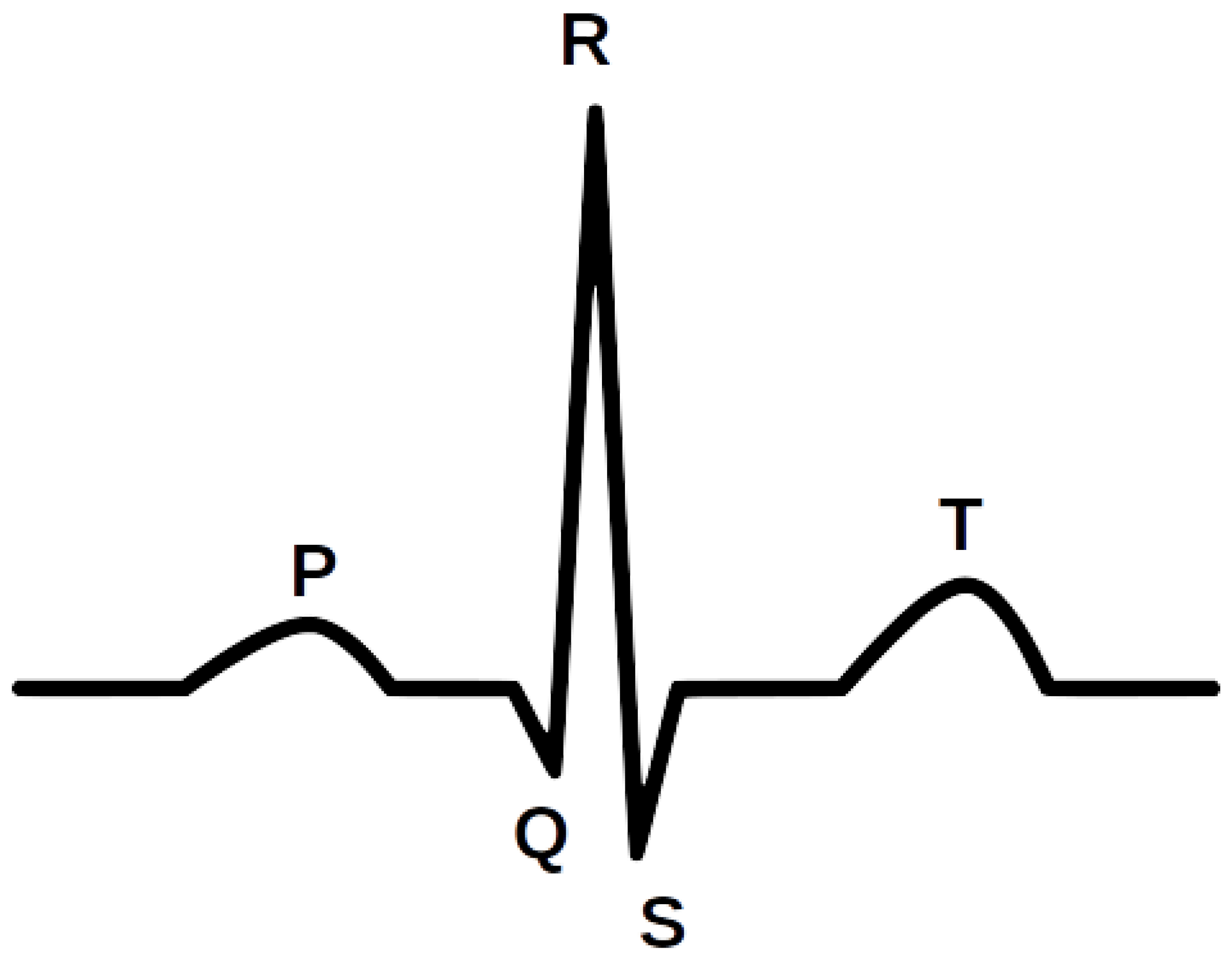
Figure 1 shows an illustrative waveform of the ECG signal. Every day, approximately 3 million ECGs are produced worldwide [
18]. ECG data contain rich information about the rate and rhythm of the heartbeat. Clinically, the ECG is analyzed over a short period using a graph of several consecutive cardiac cycles. The process begins with R-peak detection. It is usually the most visible part of the ECG that can be easily identified. The ECG reflects the depolarization of the main mass of the ventricles and refers to the maximum amplitude in the QRS complex. QRS complexes are the starting point for the analysis of the ECG signal. They serve as rhythm items and provide information about intraventricular rhythm and conduction [
19,
20].
Several methods and techniques have been used to locate the R-peak in the ECG signal, based on standard techniques such as digital filtering, wavelet transform, Fourier Transform, signal decomposition, and Hilbert Transform. However, only a few proposed works use DL methods in the literature to detect QRS complexes. One of the works in [
21] is where a 300-point Convolutional Neural Network (CNN) and clustering on the neural output are used to detect QRS complexes on the pre-processed input signal. Another method using CNN has been proposed [
22], demonstrating the reliable detection of the fetal QRS complex. The authors of the work [
23] proposed a 1-D CNN and Multi-Layer Perceptron (MLP) classifier that determines the QRS positions. Another approach was the work [
19] in which two DL models based on multi-dilated convolutional blocks were used: CNN and CRNN. Finally, this group of works includes [
24], where a stacked autoencoder deep neural network is proposed to extract the QRS complex.
Regardless of the DL methods chosen, problems are identified, including classification efficiency, the detection of undesirable results, dependence on computing power, and the high sample count. In response to these problems, a few newly published articles propose using Few-Shot Learning (FSL) to identify new concepts in medicine and fill the gap between the efficiency and the size of the training samples. FSL mimics humans’ ability to acquire knowledge from a few samples. This technique involves training a neural network to encode input data into small-sized vectors, which distances to other vectors encoding objects of the same class are smaller than to vectors representing objects from different classes. The distance between vectors is usually computed by measuring the Euclidean distance between two vectors. In addition, FSL can encode information regarding the object’s belonging to a particular class in the output vector. Because of that, the layer of neurons representing defined classes is not required, which allows the FSL network to distinguish between classes that were not seen during training, thereby enabling learning from limited samples and rapidly generalizing to new tasks, giving a different perspective on DL.
There are many areas of application of FSL methods. In the medical field, the use of FSL methods occurs in conjunction with medical images and medical signals. One of the application directions is to use the network-based FSL method to classify rare human peripheral blood leukocyte images. The proposed Siamese network by the authors of [
25] contains two identical Convolutional Neural Networks and a logistic regression network. In justifying their research, the authors point to the relationship between the number of leukocytes and various diseases, including cancer. The obtained results show that the Siamese network can overcome the scarcity and imbalance of datasets used in this research. The results are promising and give hope for addressing the issue of rare leukocyte images recognition in medicine.
Another view is the use of Few-Shot Deep Learning in medical imaging, for example, COVID-19-infected areas in Computed Tomography (CT) images. Recent studies indicate that detecting radiographic patterns on chest CT scans can provide high sensitivity and specificity in identifying COVID-19. One of the works [
26] was undertaken to investigate the efficacy of FSL in U-Net architectures, allowing for a dynamic fine-tuning of the network weights as new samples are fed into the U-Net. The obtained results confirmed the improvement of the segmentation accuracy improvement in the identification of COVID-19-infected regions. A similar approach was proposed by the authors of another study [
27], pointing to the use of FSL for the computerized diagnosis of emergencies due to coronavirus-infected pneumonia on CT images. A similar application of FSL was demonstrated by the authors of the study [
28], who undertook the classification of COVID-19 infected areas on X-rays. As part of the research, the method was tested to classify images showing unknown symptoms of COVID-19 in an environment designed to learn several samples, with prior meta-learning only on images of other diseases.
Diagnostics of disease states based on medical images using DL methods have also been applied in dermatology. The authors of the work [
29] demonstrated the possibility of using FSL for Dermatological Disease Diagnosis. Skin diseases are increasingly becoming one of the most common human diseases, contributing to dangerous cancerous changes or affecting motor disability. The proposed method is scalable to new classes and can effectively capture intra-class variability. A similar approach was used by the authors of [
30], who proposed a Few-Shot segmentation network for skin lesion segmentation, which requires only a few pixel-level annotations. The authors emphasize that the proposed method is a promising framework for Few-Shot segmentation of skin lesions. The conducted experiments show that removing the background region of the query image both accelerates the speed of network convergence and significantly improves the segmentation efficiency.
The works of other authors in medicine with the use of FSL indicate the possibility of application in creating predictive models of drug reactions based on screens of cell lines. For example, the authors’ work in [
31] applied Few-Shot machine learning to train a versatile neural network model in cell lines that can be tuned to new contexts using a few additional samples. The model quickly adapted to switching between different tissue types and shifting from cell line models to clinical contexts.
In biomedical signals, an interesting approach is to use the FSL method of Electroencephalography (EEG)-based Motor Imagery (MI) Classification. The authors of the work [
32] drew attention to an essential aspect of research on the brain–computer interface using EEG signals. In their justification, they indicated the potential of EEG in designing key technologies in both healthcare and other industries. The research proposed a two-way Few Shot network that can efficiently learn representative features of unseen subject categories and classify them with limited MI EEG data.
In the area of the ECG signal, the authors in [
33] proposed a meta-transfer-based FSL method to handle arrhythmia classification with the ECG signal in wearable devices. The results obtained by the authors indicate that the proposed method exceeds the accuracy of other comparative methods when performing various Few Shot tasks within the same training samples.
The study aimed to determine the usefulness of the FSL for ECG signal proximity-based classification. The research was conducted by training Deep Convolutional Neural Networks to recognize 2, 5, and 20 different heart disease classes. For this task, two neural networks were trained. The first one was optimized by performing FSL to classify input samples based on Euclidean distance to the defined classes’ vectors. The second one was trained to perform softmax-based classification. It serves as a basis for comparison due to its well-known effectiveness in recognizing classes established during training. This work also examines classification strategies in FSL by comparing the results obtained from proximity-based classification to training machine learning algorithms on top of optimized FSL neural networks. The tested machine learning algorithms are XGBoost, Random Forest, Decision Tree, K-Nearest Neighbors, and SVMs. The neural network proposed for this task interprets a set of QRS complexes extracted from ECG signals. The method of R-peaks labeling and QRS complexes extraction has been implemented. This procedure converts a 12-lead signal into a set of R waves by using the detection algorithms and the k-mean algorithm. The novelty of this work involves using the FSL learning style for training on known, fixed classes; its comparison with more typical, softmax-based classifications; and the evaluation of classification strategies to be employed on top of the trained FSL network.
This paper is organized as follows:
Section 2 closely describes the methods, the architectures of the artificial intelligence system, and the previously carried out data filtering, R Wave detection, and QRS extraction. Then,
Section 3 presents the result of the research. Then, the discussion is given in
Section 4. Finally,
Section 5 concludes the paper and provides a look at further studies on this topic.
4. Discussion
The Deep Neural Network trained in a Few-Shot learning (FSL) fashion for proximity-based classification provides the benefit of improved accuracy through an embedded version of online learning, allowing for continuous classification augmentation without network weight adjustments. The network’s accuracy can be improved without the additional optimization of its weights through the expansion of the classified signals dataset. Such a set is used for referential class vector computation and is essential for the correct signal classification. Cardiological professionals can improve the network by labeling the signal and increasing the number of vectors used for class vector calculation, resulting in better classification. Such a procedure does not require training of the network, which is cumbersome on production machines due to the higher computation complexity of the training network than using an already trained one. This augmentation procedure can be conducted on a CPU with low computation capabilities due to the simplicity of mean vector calculation.
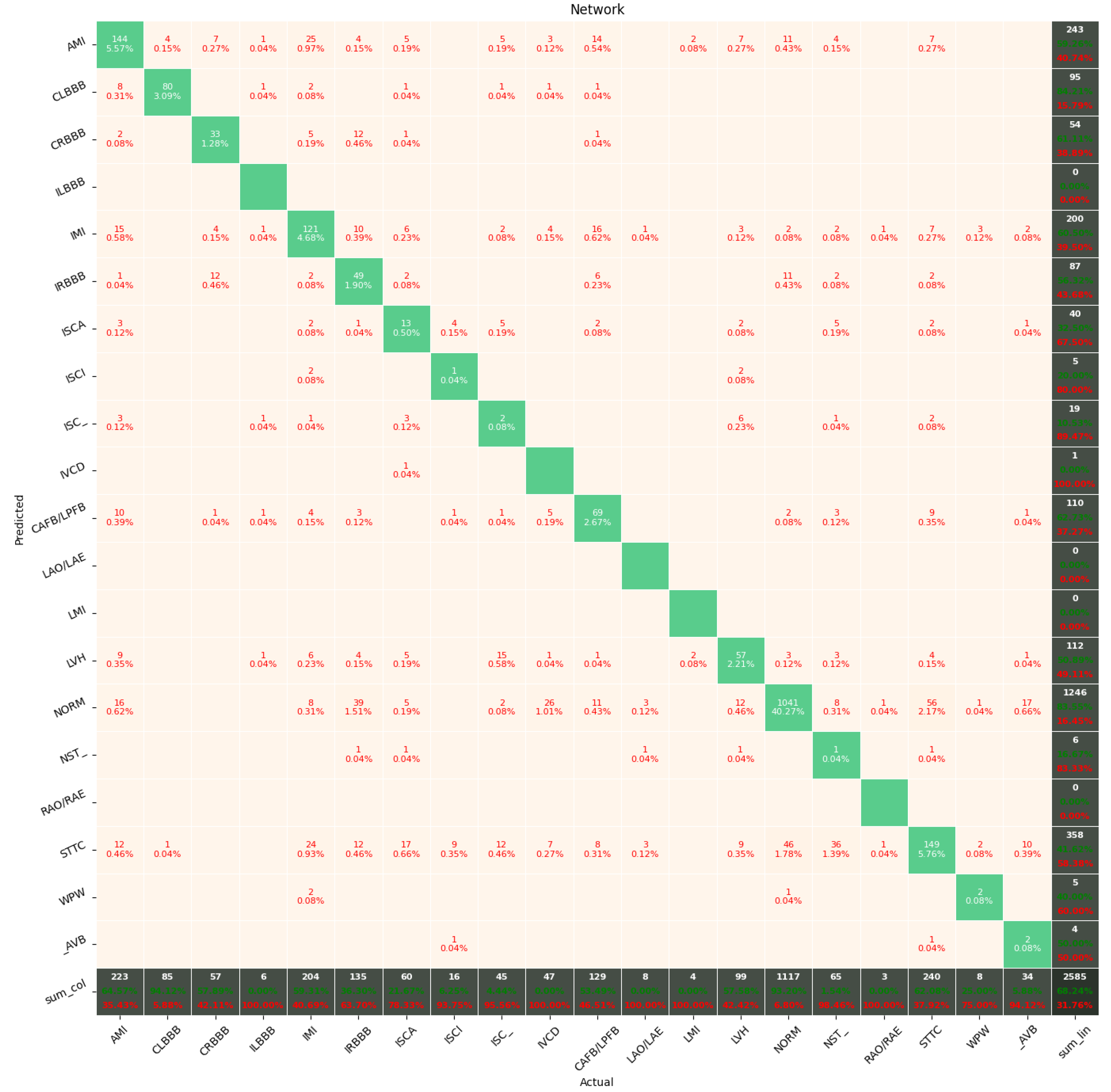
The Few-Shot Learning neural network proved to be more accurate than the softmax-based network while classifying two classes. The FSL model had higher results in both averages, maximal and minimal accuracy. However, the network proved to be less accurate on tasks involving 5- and 20-class labeling. This phenomenon is most likely a result of insufficient representation of classes with low cardinality. For example: In
Figure 11, the class “NORM” having the highest number of ECG records had the best precision and recall of all classes. The authors plan a further examination of the dataset size’s influence on the quality of prediction.
This work classified the signals processed by an FSL neural network by computing the average vector representing each class and comparing the Euclidean distance between the classified sample and all class-representing vectors. The other methods evaluated in this work for classification use network-encoded signals in small-sized vectors to train models running algorithms such as XGBoost, Random Forest, Decision Tree, K-Nearest Neighbors, and SVMs with linear, polynomial, radial basis function, and sigmoid kernels. It turned out that the most promising classification algorithm for FSL in this particular task is SVM with a radial basis function kernel. This method proved to be the most effective among all the examined FSL classification strategies and achieved better results than softmax-based classification for both two and five classes. It achieved one of the highest scores in accuracy, specificity, sensitivity, F1, and AUC among all compared models. The outcomes are promising and suggest that the hybrid neural network systems based on proximity-differentiation classification with integrated machine learning models may provide better results than the typical softmax-based state-of-the-art classification. The authors plan on conducting further research to determine whether a combination of FSL with SVM with radial basis function kernel is beneficial in other tasks or merely the case in this particular example.
The accuracy of the FSL network during the training process varies significantly more than its softmax-based counterpart. This phenomenon is depicted in
Figure 15,
Figure 16,
Figure 17,
Figure 18,
Figure 19 and
Figure 20. The softmax-based classification network reaches convergence faster and is less susceptible to the noise generated by the random selection of training data. This variance of the learning process is essential because of the commonness of early-stopping usage during network training. Typical early-stopping implemented in DL frameworks such as Keras stops the training if the evaluation score of the trained network on the validation dataset was not improved in a specific amount of time. This mechanism is important as it reduces the amount of wasted computation time and energy. However, due to the high variance of the FSL process, it is possible that controlling early-stopping based on local extremum may not be the best strategy. The results indicate that filtration of evaluation score’s signal, such as averaging, may prove beneficial. The authors plan on further examination in future works.
In previous work [
16], the best-obtained result in that research classifying sick/healthy patients (2 classes) is 89.2% accuracy. This value was increased in this research by the FSL neural network, the accuracy of which spans from 89.5% to 91.1%. As a result, even the worst performance of the studied network was better than the best in previous work. However, the results were not as promising during the classification of 5 and 20 classes. It is speculated that FSL can obtain better results for bigger datasets than Softmax-based classification, but the latter requires less training data than the former. The authors plan on conducting further research of this phenomenon.
The dataset size had almost no influence on the classification performance of the two classes. However, its impact was significant for the classification of 5 classes and even more important for the classification of 20 classes. It turns out that the more that classes are differentiated from each other, the more data are required.