Multi-Method Diagnosis of Blood Microscopic Sample for Early Detection of Acute Lymphoblastic Leukemia Based on Deep Learning and Hybrid Techniques
Abstract
:1. Introduction
- All images were optimized in both datasets by overlapping filters to obtain high-quality images
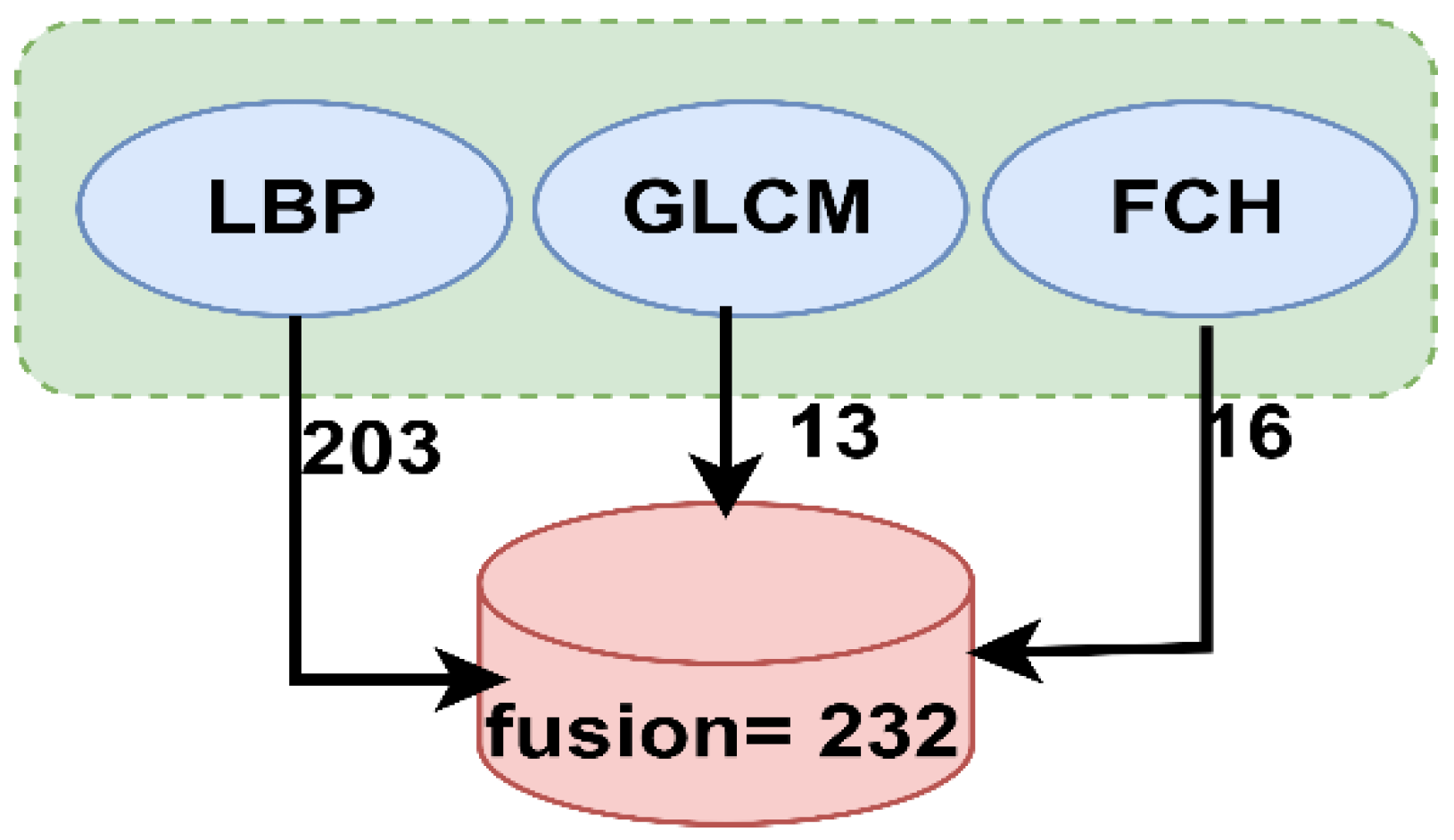
- A hybrid feature extraction technique was applied using the LBP, GLCM, and FCH algorithms, and then all the features were fused in one vector and classified using three classifiers: ANN, FFNN, and SVM
- The hybrid technique was applied between CNN models to extract deep features classified using the SVM algorithm, obtaining promising diagnostic performance results.
- Systems were developed for analyzing blood microscopy images to assist hematologists and experts in making accurate diagnostic decisions
2. Related Work
3. Materials and Methodology
3.1. Description of the Two Datasets
3.2. Enhancement of Images
3.3. Neural Networks and Machine Learning Technique
3.3.1. Adaptive Region-Growing Algorithm
- ▪
- , where m is the number of regions
- ▪
- ▪
- ▪
3.3.2. Morphological Method
3.3.3. Feature Extraction
3.3.4. Classification
Artificial Neural Network and Feed Forward Neural Network Algorithms
Support Vector Machine Algorithm
3.4. Convolutional Neural Networks (CNNs)
3.4.1. AlexNet Model
3.4.2. GoogLeNet Model
3.4.3. ResNet-18 Model
3.5. Deep Learning–Machine Learning Hybrid Techniques
4. Experimental Results
4.1. Dataset Splitting and Environment Setup
4.2. Evaluation Metrics
4.3. Results of the ANN, FFNN and SVM Algorithms
4.3.1. Performance Analysis
4.3.2. Gradient
4.3.3. Receiver Operating Characteristic (ROC)
4.3.4. Error Histogram
4.3.5. Regression
4.3.6. Confusion Matrix
4.4. Results of the Convolutional Neural Network Models
4.5. Results of the Hybrid Convolutional Neural Network Models with the Support Vector Machine Classifier
5. Discussion and Comparison with Previous Studies
6. Conclusions and Future Work
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuan, D.H.; Wu, C.C.; Su, W.Y.; Huang, N.T. A microfluidic device for simultaneous extraction of plasma, red blood cells, and on-chip white blood cell trapping. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.R.; Alagawany, M. Erythrocytes as a biological model for screening of xenobiotics toxicity. Chem. Biol. Interact. 2018, 279, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Rezatofighi, S.H.; Soltanian-Zadeh, H. Automatic recognition of five types of white blood cells in peripheral blood. Comput. Med. Imaging Graph. 2011, 35, 333–343. [Google Scholar] [CrossRef]
- Faggio, C.; Sureda, A.; Morabito, S.; Sanches-Silva, A.; Mocan, A.; Nabavi, S.F.; Nabavi, S.M. Flavonoids and platelet aggregation: A brief review. Eur. J. Pharmacol. 2017, 807, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Al-Hafiz, F.; Al-Megren, S.; Kurdi, H. Red blood cell segmentation by thresholding and Canny detector. Procedia Comput. Sci. 2018, 141, 327–334. [Google Scholar] [CrossRef]
- Rahadi, I.; Choodoung, M.; Choodoung, A. Red blood cells and white blood cells detection by image processing. J. Phys. Conf. Ser. 2020, 1539, 012025. [Google Scholar] [CrossRef]
- Van der Meijden, P.E.; Heemskerk, J.W. Platelet biology and functions: New concepts and clinical perspectives. Nat. Rev. Cardiol. 2019, 16, 166–179. [Google Scholar] [CrossRef]
- Sawyers, C.L.; Denny, C.T.; Witte, O.N. Leukemia and the disruption of normal hematopoiesis. Cell 1991, 64, 337–350. [Google Scholar] [CrossRef]
- Wehrmacher, W.H.; Messmore, H.L. Wintrobe’s Atlas of Clinical Hematology. JAMA 2007, 297, 2641–2645. [Google Scholar] [CrossRef]
- Haworth, C.; Heppleston, A.D.; Jones, P.M.; Campbell, R.H.; Evans, D.I.; Palmer, M.K. Routine bone marrow examination in the management of acute lymphoblastic leukaemia of childhood. J. Clin. Pathol. 1981, 34, 483–485. [Google Scholar] [CrossRef]
- Bain, B.J. Diagnosis from the Blood Smear. N. Engl. J. Med. 2009, 353, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Amin, J.; Sharif, M.; Anjum, M.A.; Siddiqa, A.; Kadry, S.; Nam, Y.; Raza, M. 3d semantic deep learning networks for leukemia detection. Comput. Mater. Contin. 2021, 69, 785–799. [Google Scholar] [CrossRef]
- Kumar, A.; Rawat, J.; Kumar, I.; Rashid, M.; Singh, K.U.; Al-Otaibi, Y.D.; Tariq, U. Computer-aided deep learning model for identification of lymphoblast cell using microscopic leukocyte images. Expert Syst. 2021, 29, e12894. [Google Scholar] [CrossRef]
- Shirazi, S.H.; Umar, A.I.; Naz, S.; Razzak, M.I. Efficient leukocyte segmentation and recognition in peripheral blood image. Technol. Health Care 2016, 24, 335–347. [Google Scholar] [CrossRef]
- Dhanachandra, N.; Manglem, K.; Chanu, Y.J. Image Segmentation Using K -means Clustering Algorithm and Subtractive Clustering Algorithm. Procedia Comput. Sci. 2015, 54, 764–777. [Google Scholar] [CrossRef]
- Sell, S.L.; Widen, S.G.; Prough, D.S.; Hellmich, H.L. Principal component analysis of blood microRNA datasets facilitates diagnosis of diverse diseases. PLoS ONE 2020, 15, e0234185. [Google Scholar] [CrossRef]
- Saleem, S.; Amin, J.; Sharif, M.; Anjum, M.A.; Iqbal, M.; Wang, S.H. A deep network designed for segmentation and classification of leukemia using fusion of the transfer learning models. Complex Intell. Syst. 2021, 1, 1–16. [Google Scholar] [CrossRef]
- Mirmohammadi, P.; Ameri, M.; Shalbaf, A. Recognition of acute lymphoblastic leukemia and lymphocytes cell subtypes in microscopic images using random forest classifier. Phys. Eng. Sci. Med. 2021, 44, 433–444. [Google Scholar] [CrossRef]
- Genovese, A.; Hosseini, M.S.; Piuri, V.; Plataniotis, K.N.; Scotti, F. Acute Lymphoblastic Leukemia Detection Based on Adaptive Unsharpening and Deep Learning. In Proceedings of the ICASSP 2021-2021 IEEE International Conference on Acoustics Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 1205–1209. [Google Scholar] [CrossRef]
- Mohapatra, S.; Patra, D.; Satpathy, S. An ensemble classifier system for early diagnosis of acute lymphoblastic leukemia in blood microscopic images. Neural Comput. Appl. 2014, 24, 1887–1904. [Google Scholar] [CrossRef]
- Mahmood, N.; Shahid, S.; Bakhshi, T.; Riaz, S.; Ghufran, H.; Yaqoob, M. Identification of significant risks in pediatric acute lymphoblastic leukemia (all) through machine learning (mL) approach. Med Biol. Eng. Comput. 2020, 58, 2631–2640. [Google Scholar] [CrossRef]
- Alrefai, N. Ensemble Machine Learning for Leukemia Cancer Diagnosis based on Microarray Datasets. Int. J. Appl. Eng. Res. 2019, 14, 4077–4084. Available online: http://www.ripublication.com (accessed on October 2021).
- Mandal, S.; Daivajna, V.; Rajagopalan, V. Machine learning based system for automatic detection of leukemia cancer cell. In Proceedings of the 2019 IEEE 16th India Council International Conference (INDICON), Rajkot, India, 13–15 December 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Naz, I.; Muhammad, N.; Yasmin, M.; Sharif, M.; Shah, J.H.; Fernandes, S.L. Robust discrimination of leukocytes protuberant types for early diagnosis of leukemia. J. Mech. Med. Biol. 2019, 19, 1950055. [Google Scholar] [CrossRef]
- Roy, R.M.; Ameer, P.M. Segmentation of leukocyte by semantic segmentation model: A deep learning approach. Biomed. Signal Process. Control. 2021, 65, 102385. [Google Scholar] [CrossRef]
- Schouten, J.P.; Matek, C.; Jacobs, L.F.; Buck, M.C.; Bošnački, D.; Marr, C. Tens of images can suffice to train neural networks for malignant leukocyte detection. Sci. Rep. 2021, 11, 1–8. [Google Scholar] [CrossRef]
- Labati, R.D.; Piuri, V.; Scotti, F. All-IDB: The acute lymphoblastic leukemia image database for image processing. In Proceedings of the 18th IEEE International Conference on Image Processing IEEE, Brussels, Belgium, 11–14 September 2011; pp. 2045–2048. [Google Scholar] [CrossRef]
- Basima, C.T.; Panicker, J.R. Enhanced leucocyte classification for leukaemia detection. In Proceedings of the 2016 International Conference on Information Science (ICIS) IEEE, Kochi, India, 2–13 August 2016; pp. 65–71. [Google Scholar] [CrossRef]
- Abunadi, I.; Senan, E.M. Deep Learning and Machine Learning Techniques of Diagnosis Dermoscopy Images for Early Detection of Skin Diseases. Electronics 2021, 10, 3158. [Google Scholar] [CrossRef]
- Mishra, S.; Majhi, B.; Sa, P.K.; Sharma, L. Gray level co-occurrence matrix and random forest based acute lymphoblastic leukemia detection. Biomed. Signal Process. Control. 2017, 33, 272–280. [Google Scholar] [CrossRef]
- Senan, E.M.; Jadhav, M.E. Techniques for the detection of skin lesions in PH2 dermoscopy images using local binary pattern (LBP). In Recent Trends in Image Processing and Pattern Recognition; RTIP2R 2020; Communications in Computer and Information Science; Santosh, K.C., Gawali, B., Eds.; Springer: Singapore, 2021; Volume 1381, pp. 14–25. [Google Scholar] [CrossRef]
- Senan, E.M.; Alzahrani, A.; Alzahrani, M.Y.; Alsharif, N.; Aldhyani, T.H. Automated Diagnosis of Chest X-Ray for Early Detection of COVID-19 Disease. Comput. Math. Methods Med. 2021, 2021, 6919483. [Google Scholar] [CrossRef]
- Abbas, Z.; Rehman, M.U.; Najam, S.; Rizvi, S.D. An efficient gray-level co-occurrence matrix (GLCM) based approach towards classification of skin lesion. In Proceedings of the 2019 Amity International Conference on Artificial Intelligence (AICAI) IEEE, Dubai, United Arab Emirates, 4–6 February 2019; pp. 317–320. [Google Scholar] [CrossRef]
- Mishra, S.; Majhi, B.; Sa, P.K. Texture feature based classification on microscopic blood smear for acute lymphoblastic leukemia detection. Biomed. Signal Process. Control. 2019, 47, 303–311. [Google Scholar] [CrossRef]
- Deng, L.; Yu, D. Deep Learning. Found. Trends Signal Process. 2014, 7, 197–387. [Google Scholar] [CrossRef]
- Yoshua, B. Learning Deep Architectures for AI. 2009, p. 127. Available online: https://books.google.com/books/about/Learning_Deep_Architectures_for_AI.html?id=cq5ewg7FniMC (accessed on 15 December 2021).
- Senan, E.M.; Abunadi, I.; Jadhav, M.E.; Fati, S.M. Score and Correlation Coefficient-Based Feature Selection for Predicting Heart Failure Diagnosis by Using Machine Learning Algorithms. Comput. Math. Methods Med. 2021, 2021, 85003. [Google Scholar] [CrossRef]
- Hmoud Al-Adhaileh, M.; Mohammed Senan, E.; Alsaade, W.; Aldhyani, T.H.; Alsharif, N.; Abdullah Alqarni, A.; Jadhav, M.E. Deep Learning Algorithms for Detection and Classification of Gastrointestinal Diseases. Complexity 2021, 2021, 6170416. [Google Scholar] [CrossRef]
- Ahmed, I.A.; Senan, E.M.; Rassem, T.H.; Ali, M.A.; Shatnawi, H.S.A.; Alwazer, S.M.; Alshahrani, M. Eye Tracking-Based Diagnosis and Early Detection of Autism Spectrum Disorder Using Machine Learning and Deep Learning Techniques. Electronics 2022, 11, 530. [Google Scholar] [CrossRef]
- Mohammed, B.A.; Senan, E.M.; Rassem, T.H.; Makbol, N.M.; Alanazi, A.A.; Al-Mekhlafi, Z.G.; Almurayziq, T.S.; Ghaleb, F.A. Multi-Method Analysis of Medical Records and MRI Images for Early Diagnosis of Dementia and Alzheimer’s Disease Based on Deep Learning and Hybrid Methods. Electronics 2021, 10, 2860. [Google Scholar] [CrossRef]
- Senan, E.M.; Alsaade, F.W.; Al-mashhadani, M.I.A.; Theyazn, H.H.; Al-Adhaileh, M.H. Classification of histopathological images for early detection of breast cancer using deep learning. J. Appl. Sci. Eng. 2021, 24, 323–329. [Google Scholar] [CrossRef]
- Ramaneswaran, S.; Srinivasan, K.; Vincent, P.M.; Chang, C.Y. Hybrid Inception v3 XGBoost Model for Acute Lymphoblastic Leukemia Classification. Comput. Math. Methods Med. 2021, 2021, 2577375. Available online: https://www.hindawi.com/journals/cmmm/2021/2577375/ (accessed on October 2021). [CrossRef]
- Shafique, S.; Tehsin, S. Acute lymphoblastic leukemia detection and classification of its subtypes using pretrained deep convolutional neural networks. Technol. Cancer Res. Treat. 2018, 17, 1–7. [Google Scholar] [CrossRef]
- Putzu, L.; Caocci, G.; Di Ruberto, C. Leucocyte classification for leukaemia detection using image processing techniques. Artif. Intell. Med. 2014, 62, 179–191. [Google Scholar] [CrossRef]
- Amin, J.; Sharif, M.; Anjum, M.A. An Integrated Design Based on Dual Thresholding and Features Optimization for White Blood Cells Detection. IEEE Access 2021, 9, 151421–151433. [Google Scholar] [CrossRef]
- Shafique, S.; Tehsin, S.; Anas, S.; Masud, F. Computer-assisted acute lymphoblastic leukemia detection and diagnosis. In Proceedings of the 2019 2nd International Conference on Communication, Computing and Digital systems, Islamabad, Pakistan, 6–7 March 2019; pp. 184–189. [Google Scholar] [CrossRef]





























| Dataset | ALL_IDB1 | ALL_IDB2 | ||||
|---|---|---|---|---|---|---|
| Phase | 80% for training and validation (80:20%) | 20% for testing | 80% for training and validation (80:20%) | 20% for testing | ||
| Classes | Training (80%) | validation (20%) | Training (80%) | validation (20%) | ||
| Leukemia | 31 | 8 | 10 | 83 | 21 | 26 |
| Normal | 38 | 9 | 12 | 83 | 21 | 26 |
| Dataset | ALL_IDB1 | ALL_IDB1 | ||||
|---|---|---|---|---|---|---|
| Classifiers | ANN | FFNN | SVM | ANN | FFNN | SVM |
| Accuracy % | 94.4 | 100 | 90.91 | 100 | 100 | 98.11 |
| Precision % | 100 | 100 | 100 | 100 | 100 | 95.83 |
| Sensitivity % | 91.55 | 100 | 84.62 | 100 | 100 | 100 |
| Specificity % | 91.55 | 100 | 100 | 100 | 100 | 96.67 |
| AUC % | 94.52 | 100 | 91.99 | 99.21 | 100 | 97.86 |
| Dataset | ALL_IDB1 | ALL_IDB2 | ||
|---|---|---|---|---|
| Phase | During the training phase | During the training phase | ||
| Classes | Leukemia | Normal | Leukemia | Normal |
| No images before augmentation | 39 | 47 | 104 | 104 |
| No images after augmentation | 780 | 799 | 1040 | 1040 |
| Options | AlexNet | GoogleNet | ResNet-18 |
|---|---|---|---|
| training Options | adam | adam | adam |
| Mini Batch Size | 130 | 20 | 15 |
| Max Epochs | 10 | 5 | 8 |
| Initial Learn Rate | 0.0001 | 0.0003 | 0.0001 |
| Validation Frequency | 50 | 3 | 5 |
| Execution Environment | 4 GB GPU | 4 GB GPU | 4 GB GPU |
| Dataset | ALL_IDB1 | ALL_IDB2 | ||||
|---|---|---|---|---|---|---|
| Measure | AlexNet | GoogLeNet | ResNet-18 | AlexNet | GoogLeNet | ResNet-18 |
| Accuracy % | 100 | 100 | 100 | 94.2 | 100 | 97.44 |
| Precision % | 100 | 100 | 100 | 92.3 | 100 | 97.4 |
| Sensitivity % | 100 | 100 | 100 | 96.2 | 100 | 97.4 |
| Sepecificy % | 100 | 100 | 100 | 94.5 | 100 | 97.4 |
| AUC % | 100 | 100 | 100 | 99.26 | 100 | 99.93 |
| Dataset | ALL_IDB1 | ALL_IDB2 | ||||
|---|---|---|---|---|---|---|
| Measure | AlexNet + SVM | GoogLeNet + SVM | ResNet-18 + SVM | AlexNet + SVM | GoogLeNet + SVM | ResNet-18 + SVM |
| Accuracy % | 100 | 95.5 | 100 | 96.2 | 98.1 | 100 |
| Precision % | 100 | 95.45 | 100 | 96.2 | 98 | 100 |
| Sensitivity % | 100 | 100 | 100 | 96.2 | 96.2 | 100 |
| Specificity % | 100 | 91.7 | 100 | 96.2 | 100 | 100 |
| AUC % | 100 | 99.34 | 100 | 98.56 | 99.87 | 100 |
| Dataset | Diseases | Neural Networks and Machine Learning | Deep Learning | Hybrid | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ANN | FFNN | SVM | Alex-Net | Google-Net | Res-Net-18 | AlexNett + SVM | GoogleNet + SVM | ResNet-18 + SVM | ||
| ALL_IDB1 | Leukemia | 87.4 | 100 | 84.62 | 100 | 100 | 100 | 100 | 100 | 100 |
| Normal | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 91.7 | 100 | |
| ALL_IDB2 | Leukemia | 100 | 100 | 100 | 96.2 | 100 | 97.4 | 96.2 | 96.2 | 100 |
| Normal | 100 | 100 | 95.83 | 92.3 | 100 | 97.4 | 96.2 | 100 | 100 | |
| Previous Studies | Accuracy % | Precision % | Sensitivity % | Specificity % | AUC % |
|---|---|---|---|---|---|
| S. Ramaneswaran.; et al. [42] | 89.40 | 90.10 | 88.90 | - | 83.20 |
| Sarmad Shafique.; et al. [43] | 93.33 | 94.01 | 94.82 | 98.4 | - |
| Lorenzo Putzu.; et al. [44] | 93.00 | - | 98.00 | - | - |
| Javaria Amin.; et al. [45] | 91.59 | - | 93.33 | 87.50 | 97.46 |
| Sarmad Shafique.; et al. [46] | 93.70 | - | 92.00 | 91 | - |
| Proposed model | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abunadi, I.; Senan, E.M. Multi-Method Diagnosis of Blood Microscopic Sample for Early Detection of Acute Lymphoblastic Leukemia Based on Deep Learning and Hybrid Techniques. Sensors 2022, 22, 1629. https://doi.org/10.3390/s22041629
Abunadi I, Senan EM. Multi-Method Diagnosis of Blood Microscopic Sample for Early Detection of Acute Lymphoblastic Leukemia Based on Deep Learning and Hybrid Techniques. Sensors. 2022; 22(4):1629. https://doi.org/10.3390/s22041629
Chicago/Turabian StyleAbunadi, Ibrahim, and Ebrahim Mohammed Senan. 2022. "Multi-Method Diagnosis of Blood Microscopic Sample for Early Detection of Acute Lymphoblastic Leukemia Based on Deep Learning and Hybrid Techniques" Sensors 22, no. 4: 1629. https://doi.org/10.3390/s22041629
APA StyleAbunadi, I., & Senan, E. M. (2022). Multi-Method Diagnosis of Blood Microscopic Sample for Early Detection of Acute Lymphoblastic Leukemia Based on Deep Learning and Hybrid Techniques. Sensors, 22(4), 1629. https://doi.org/10.3390/s22041629







