Classifying Muscle States with One-Dimensional Radio-Frequency Signals from Single Element Ultrasound Transducers
Abstract
:1. Introduction
1.1. Related Work
1.2. Our Contribution
2. Materials and Methods
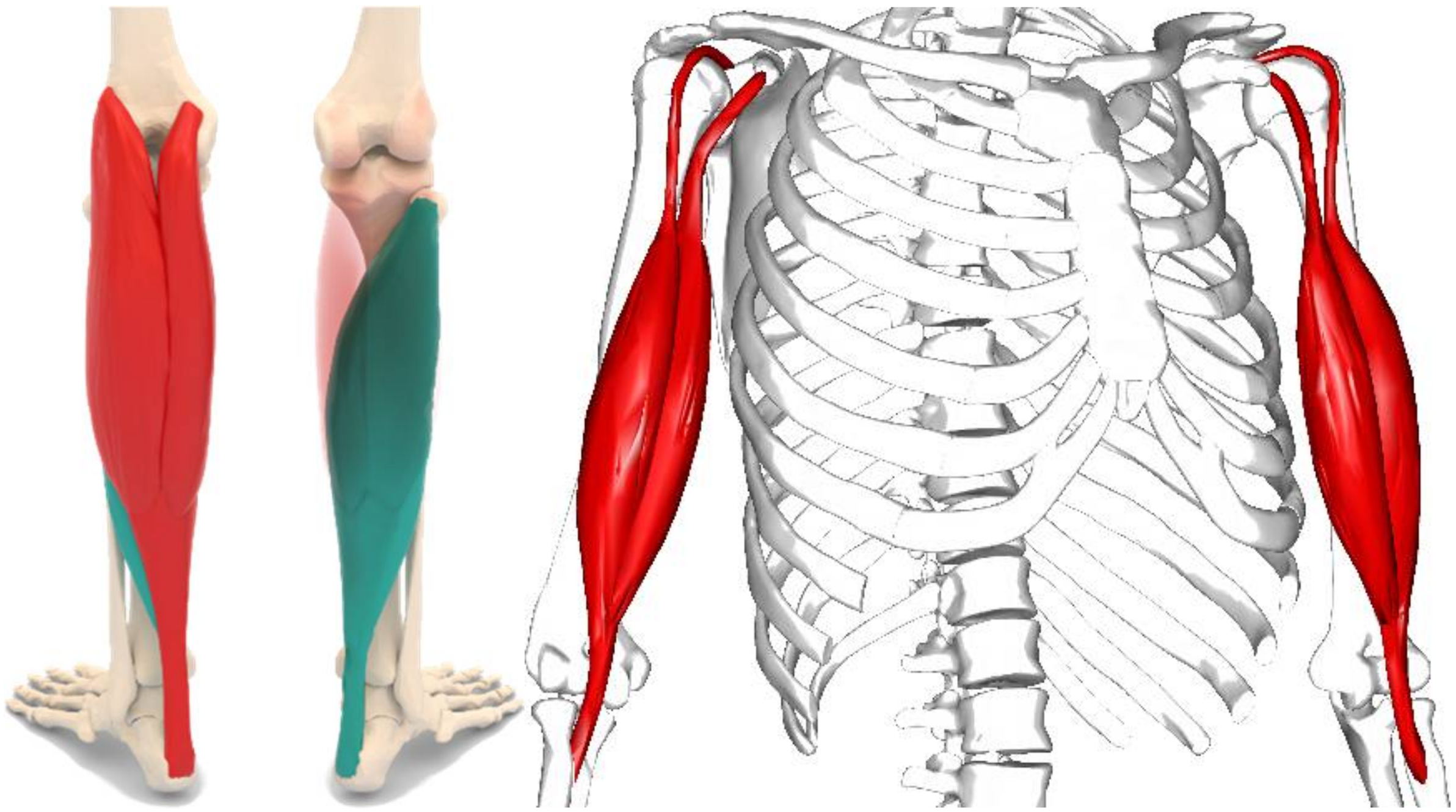
2.1. Materials
2.2. Methods
2.3. Experimental Setup
2.4. Evaluation
3. Results
3.1. Muscle Contraction Signals Classifications
3.1.1. T-Distributed Stochastic Neighbor Embedding
3.1.2. Machine Learning
3.2. Muscle Fatigue Signals Classifications
3.2.1. T-Distributed Stochastic Neighbor Embedding on Signals of Study One
3.2.2. T-Distributed Stochastic Neighbor Embedding on Signals of Study Two
3.2.3. Machine Learning
4. Discussion
4.1. Muscle Contraction Signals Classifications
4.2. Muscle Fatigue Signals Classification
4.3. Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Dataset ID | Subject ID | Gender | Unique Transducer Position | # A-Scans | A-Scans/s |
|---|---|---|---|---|---|
| 1 | 01 | male | no | 3000 | 54.71 |
| 2 | 01 | male | no | 3000 | 50.55 |
| 3 | 02 | male | no | 1000 | 55.32 |
| 4 | 02 | male | no | 1000 | 52.06 |
| 5 | 01 | male | yes | 6000 | 56.17 |
| 6 | 01 | male | yes | 50,000 | 33.95 |
| 7 | 03 | female | yes | 10,000 | 48.56 |
| 8 | 03 | female | yes | 8872 | 56.17 |
| 9 | 04 | male | yes | 10,000 | 48.65 |
| 10 | 04 | male | yes | 10,000 | 38.98 |
| 11 | 05 | male | yes | 10,000 | 40.82 |
| 12 | 06 | male | yes | 10,000 | 45.33 |
| 13 | 07 | male | no | 10,000 | 58.73 |
| 14 | 07 | male | no | 10,000 | 59.55 |
| 15 | 01 | male | yes | 10,000 | 59.97 |
| 16 | 01 | male | yes | 10,000 | 60.33 |
| 17 | 01 | male | yes | 10,000 | 60.19 |
| 18 | 08 | male | no | 10,000 | 47.58 |
| 19 | 08 | male | no | 10,000 | 47.48 |
| 20 | 08 | male | yes | 10,000 | 54.44 |
| 21 | 08 | male | yes | 10,000 | 44.88 |
Appendix B
| Subject ID | Gender | # Datasets | # A-Scans | Max. Lifted Weight [kg] |
|---|---|---|---|---|
| 01 | female | 4 | 1390 | 5.0 |
| 02 | female | 3 | 1043 | 2.5 |
| 03 | male | 2 | 696 | 2.5 |
| 04 | male | 2 | 685 | 2.5 |
| 05 | male | 2 | 695 | 7.5 |
| 06 | male | 4 | 1386 | 7.5 |
| 07 | female | 4 | 1367 | 5.0 |
| 08 | male | 3 | 1044 | 5.0 |
| 09 | male | 10 | 3453 | 7.5 |
| 10 | female | 3 | 1044 | 5.0 |
| 11 | female | 2 | 696 | 5.0 |
| 12 | male | 2 | 695 | 5.0 |
| 13 | male | 3 | 1035 | 7.5 |
| 14 | female | 2 | 695 | 5.0 |
| 15 | male | 2 | 666 | 5.0 |
| 16 | male | 2 | 672 | 5.0 |
| 17 | male | 1 | 348 | 7.5 |
| 18 | male | 3 | 1035 | 5.0 |
| 19 | male | 1 | 342 | 7.5 |
| 20 | male | 1 | 345 | 7.5 |
| 21 | female | 1 | 345 | 2.5 |
| Subject ID | Gender | # Datasets | # A-Scans | Max. Lifted Weight [kg] |
|---|---|---|---|---|
| 09 | male | 42 | 13,160 | 7.5 |
References
- Lukowicz, P.; Hanser, F.; Szubski, C.; Schobersberger, W. Detecting and interpreting muscle activity with wearable force sensors. In Proceedings of the International Conference on Pervasive Computing, Pisa, Italy, 13–17 March 2006; pp. 101–116. [Google Scholar]
- Mokaya, F.; Lucas, R.; Noh, H.Y.; Zhang, P. Burnout: A wearable system for unobtrusive skeletal muscle fatigue estimation. In Proceedings of the 2016 15th ACM/IEEE International Conference on Information Processing in Sensor Networks (IPSN), Vienna, Austria, 11–14 April 2016; pp. 1–12. [Google Scholar]
- Islam, M.A.; Sundaraj, K.; Ahmad, R.B.; Ahamed, N.U. Mechanomyogram for muscle function assessment: A review. PLoS ONE 2013, 8, e58902. [Google Scholar] [CrossRef] [PubMed]
- Woodward, R.B.; Stokes, M.J.; Shefelbine, S.J.; Vaidyanathan, R. Segmenting mechanomyography measures of muscle activity phases using inertial data. Sci. Rep. 2019, 9, 5569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, M.H.; Ahn, S.J.; Lee, J.W.; Rhee, M.H.; Chae, D.; Kim, J.; Shin, M.J. Validity and reliability of the newly developed surface electromyography device for measuring muscle activity during voluntary isometric contraction. Comput. Math. Methods Med. 2018, 2018, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toro, S.F.D.; Santos-Cuadros, S.; Olmeda, E.; Álvarez-Caldas, C.; Díaz, V.; San Román, J.L. Is the use of a low-cost sEMG sensor valid to measure muscle fatigue? Sensors 2019, 19, 3204. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Sundholm, M.; Cheng, J.; Cruz, H.; Lukowicz, P. Measuring muscle activities during gym exercises with textile pressure mapping sensors. Pervasive Mob. Comput. 2017, 38, 331–345. [Google Scholar] [CrossRef]
- Gibas, C.; Grünewald, A.; Wunderlich, H.W.; Marx, P.; Brück, R. A wearable EIT system for detection of muscular activity in the extremities. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 2496–2499. [Google Scholar]
- Leitner, C.; Hager, P.A.; Penasso, H.; Tilp, M.; Benini, L.; Peham, C.; Baumgartner, C. Ultrasound as a tool to study muscle–tendon functions during locomotion: A systematic review of applications. Sensors 2019, 19, 4316. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.Z.H.; Ling, Y.T.; Shea, Q.T.K.; Wang, L.K.; Wang, X.Y.; Zheng, Y.P. Towards wearable comprehensive capture and analysis of skeletal muscle activity during human locomotion. Sensors 2019, 19, 195. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.Y.; Zheng, Y.P.; Huang, Q.H.; Chen, X. Dynamic monitoring of forearm muscles using one-dimensional sonomyography system. J. Rehabil. Res. Dev. 2008, 45, 187–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.Y.; Zheng, Y.P.; Huang, Q.H.; Chen, X.; He, J.F.; Chan, H.L.W. Performances of one-dimensional sonomyography and surface electromyography in tracking guided patterns of wrist extension. Ultrasound Med. Biol. 2009, 35, 894–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Zheng, Y.P.; Guo, J.Y.; Shi, J. Sonomyography (SMG) control for powered prosthetic hand: A study with normal subjects. Ultrasound Med. Biol. 2010, 36, 1076–1088. [Google Scholar] [CrossRef]
- Guo, J.Y.; Zheng, Y.P.; Xie, H.B.; Koo, T.K. Towards the application of one-dimensional sonomyography for powered upper-limb prosthetic control using machine learning models. Prosthet. Orthot. Int. 2013, 37, 43–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Li, Y.; Liu, H. Muscle fatigue assessment using one-channel single-element ultrasound transducer. In Proceedings of the 2017 8th International IEEE/EMBS Conference on Neural Engineering (NER), Shanghai, China, 25–28 May 2017; pp. 122–125. [Google Scholar]
- He, J.; Luo, H.; Jia, J.; Yeow, J.T.; Jiang, N. Wrist and finger gesture recognition with single-element ultrasound signals: A comparison with single-channel surface electromyogram. IEEE Trans. Biomed. Eng. 2018, 66, 1277–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Liu, J.; Zeng, J.; Li, K.; Liu, H. Bio-signal based elbow angle and torque simultaneous prediction during isokinetic contraction. Sci. China Technol. Sci. 2019, 62, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Bielemann, R.M.; Gonzalez, M.C.; Barbosa-Silva, T.G.; Orlandi, S.P.; Xavier, M.O.; Bergmann, R.B.; Assunção, M.C.F. Estimation of body fat in adults using a portable A-mode ultrasound. Nutrition 2016, 32, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Kuehne, T.E.; Yitzchaki, N.; Jessee, M.B.; Graves, B.S.; Buckner, S.L. A comparison of acute changes in muscle thickness between A-mode and B-mode ultrasound. Physiol. Meas. 2019, 40, 115004. [Google Scholar] [CrossRef]
- Yan, J.; Yang, X.; Chen, Z.; Liu, H. Dynamically characterizing skeletal muscles via acoustic non-linearity parameter: In vivo assessment for upper arms. Ultrasound Med. Biol. 2020, 46, 315–324. [Google Scholar] [CrossRef] [PubMed]
- AlMohimeed, I.; Ono, Y. Ultrasound measurement of skeletal muscle contractile parameters using flexible and wearable single-element ultrasonic sensor. Sensors 2020, 20, 3616. [Google Scholar] [CrossRef] [PubMed]
- Brausch, L.; Hewener, H.; Lukowicz, P. Towards a wearable low-cost ultrasound device for classification of muscle activity and muscle fatigue. In Proceedings of the 23rd International Symposium on Wearable Computers, London, UK, 9–13 September 2019; pp. 20–22. [Google Scholar]
- Brausch, L.; Hewener, H. Classifying muscle states with ultrasonic single element transducer data using machine learning strategies. Proc. Meet. Acoust. 2019, 38, 022001. [Google Scholar]
- Mitsuhashi, N.; Fujieda, K.; Tamura, T.; Kawamoto, S.; Takagi, T.; Okubo, K. BodyParts3D: 3D structure database for anatomical concepts. Nucleic Acids Res. 2009, 37, D782–D785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenninger, M.; Bayerl, S.P.; Schmidt, J.; Riedhammer, K. Timage—A robust time series classification pipeline. In International Conference on Artificial Neural Networks; Springer: Berlin/Heidelberg, Germany, 2019; pp. 450–461. [Google Scholar]
- Berndt, D.J.; Clifford, J. Using dynamic time warping to find patterns in time series. In Proceedings of the KDD Workshop, Seattle, WA, USA, 31 July–1 August 1994; Volume 10, pp. 359–370. [Google Scholar]
- Wang, Z.; Yan, W.; Oates, T. Time series classification from scratch with deep neural networks: A strong baseline. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 1578–1585. [Google Scholar]
- Ismail Fawaz, H.; Forestier, G.; Weber, J.; Idoumghar, L.; Muller, P.A. Deep learning for time series classification: A review. Data Min. Knowl. Discov. 2019, 33, 917–963. [Google Scholar] [CrossRef] [Green Version]
- Schwenker, F.; Dietrich, C.; Kestler, H.A.; Riede, K.; Palm, G. Radial basis function neural networks and temporal fusion for the classification of bioacoustic time series. Neurocomputing 2003, 51, 265–275. [Google Scholar] [CrossRef]
- Vidnerova, P. RBF-Keras: An RBF Layer for Keras Library. 2020. Available online: https://github.com/PetraVidnerova/rbf_keras (accessed on 4 April 2022).
- Dempster, A.; Petitjean, F.; Webb, G.I. ROCKET: Exceptionally fast and accurate time series classification using random convolutional kernels. Data Min. Knowl. Discov. 2020, 34, 1454–1495. [Google Scholar] [CrossRef]
- Dempster, A.; Schmidt, D.F.; Webb, G.I. Minirocket: A very fast (almost) deterministic transform for time series classification. In Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery Data Mining, Singapore, 14–18 August 2021; pp. 248–257. [Google Scholar]
- Tan, C.W.; Dempster, A.; Bergmeir, C.; Webb, G.I. MultiRocket: Effective summary statistics for convolutional outputs in time series classification. arXiv 2021, arXiv:2102.00457. [Google Scholar]
- Liu, M.; Ren, S.; Ma, S.; Jiao, J.; Chen, Y.; Wang, Z.; Song, W. Gated Transformer Networks for Multivariate Time Series Classification. arXiv 2021, arXiv:2103.14438. [Google Scholar]
- Allam Jr, T.; McEwen, J.D. Paying Attention to Astronomical Transients: Photometric Classification with the Time-Series Transformer. arXiv 2021, arXiv:2105.06178. [Google Scholar]
- Prokhorenkova, L.; Gusev, G.; Vorobev, A.; Dorogush, A.V.; Gulin, A. CatBoost: Unbiased boosting with categorical features. Adv. Neural Inf. Processing Syst. 2018, 31, 1–11. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Processing Syst. 2017, 30, 3149–3157. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Chollet, F.; Zhu, Q.S.; Gardener, T.; Rahman, F.; Lee, T.; De Marmiesse, G.; Zabluda, O.; ChentaMS; Watson, M.; Santana, E.; et al. Keras. GitHub. 2015. Available online: https://github.com/fchollet/keras (accessed on 4 April 2022).
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Meert, W.; Group, D.R. DTAIDistance. 2022. Available online: https://dtaidistance.readthedocs.io/ (accessed on 4 April 2022).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Cunningham, J.P.; Ghahramani, Z. Linear dimensionality reduction: Survey, insights, and generalizations. J. Mach. Learn. Res. 2015, 16, 2859–2900. [Google Scholar]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008. 9, 2579–2605.
- Gandevia, S.C. Spinal and supraspinal factors in human muscle fatigue. Physiol. Rev. 2001, 81, 1725–1789. [Google Scholar] [CrossRef] [PubMed]
- Brausch, L.; Hewener, H.; Lukowicz, P. Muscle Contraction A-Scan data annotated by volunteers. 2019. Available online: https://www.openml.org/d/41971 (accessed on 4 April 2022).
- Brausch, L.; Hewener, H.; Lukowicz, P. Muscle Fatigue A-Scan data of 21 volunteers (study 1/2). 2021. Available online: https://www.openml.org/d/43075 (accessed on 4 April 2022).
- Brausch, L.; Hewener, H.; Lukowicz, P. Muscle Contraction A-Scan data of a single volunteer (study 2/2). 2021. Available online: https://www.openml.org/d/43076 (accessed on 4 April 2022).
- Zhou, G.Q.; Zheng, Y.P.; Zhou, P. Measurement of gender differences of gastrocnemius muscle and tendon using sonomyography during calf raises: A pilot study. BioMed Res. Int. 2017, 2017, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; O’Dell, M.; He, W.; Du, L.J.; Li, P.C.; Gao, J. Ultrasound shear wave elastography in the assessment of passive biceps brachii muscle stiffness: Influences of sex and elbow position. Clin. Imaging 2017, 45, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Barandas, M.; Folgado, D.; Fernandes, L.; Santos, S.; Abreu, M.; Bota, P.; Liu, H.; Schultz, T.; Gamboa, H. TSFEL: Time series feature extraction library. SoftwareX 2020, 11, 100456. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Fawaz, H.I.; Forestier, G.; Weber, J.; Idoumghar, L.; Muller, P.A. Transfer learning for time series classification. In Proceedings of the 2018 IEEE International Conference On Big Data (Big Data), Seattle, WA, USA, 10–13 December 2018; pp. 1367–1376. [Google Scholar]
- Xia, W.; Zhou, Y.; Yang, X.; He, K.; Liu, H. Toward portable hybrid surface electromyography/a-mode ultrasound sensing for human–machine interface. IEEE Sens. J. 2019, 19, 5219–5228. [Google Scholar] [CrossRef]













| Subject ID | Gender | # A-Scans | # Datasets | # Squats |
|---|---|---|---|---|
| 1 | Male | 92,000 | 7 | 154 |
| 2 | Male | 2000 | 2 | 8 |
| 3 | Female | 18,872 | 2 | 35 |
| 4 | Male | 20,000 | 2 | 49 |
| 5 | Male | 10,000 | 1 | 27 |
| 6 | Male | 10,000 | 1 | 21 |
| 7 | Male | 20,000 | 2 | 88 |
| 8 | Male | 40,000 | 4 | 133 |
| Subject ID | Gender | # A-Scans | # Datasets | Max. Weight [kg] |
|---|---|---|---|---|
| 01 | Female | 18,283 | 4 | 5.0 |
| 02 | Female | 15,155 | 3 | 2.5 |
| 03 | Male | 16,161 | 2 | 2.5 |
| 04 | Male | 18,863 | 2 | 2.5 |
| 05 | Male | 4302 | 2 | 7.5 |
| 06 | Male | 27,112 | 4 | 7.5 |
| 07 | Female | 13,585 | 4 | 5.0 |
| 08 | Male | 8809 | 3 | 5.0 |
| 09 | Male | 109,964 | 51 | 7.5 |
| 10 | Female | 7967 | 3 | 5.0 |
| 11 | Female | 3326 | 2 | 5.0 |
| 12 | Male | 4349 | 2 | 5.0 |
| 13 | Male | 14,691 | 3 | 7.5 |
| 14 | Female | 6950 | 2 | 5.0 |
| 15 | Male | 12,817 | 2 | 5.0 |
| 16 | Male | 10,218 | 2 | 5.0 |
| 17 | Male | 3792 | 1 | 7.5 |
| 18 | Male | 14,005 | 3 | 5.0 |
| 19 | Male | 5236 | 2 | 7.5 |
| 20 | Male | 4635 | 2 | 7.5 |
| 21 | Female | 11,817 | 2 | 2.5 |
| Evaluation Mode | Study | Signals Taken from Study [%] |
|---|---|---|
| Leave-one-out cross-validation (LOOCV) on all signals | 1 | 100 |
| LOOCV on signals from dominant arm only | 1 | 52.48 |
| LOOCV on signals from non-dominant arm only | 1 | 47.52 |
| LOOCV on signals from female subjects only | 1 | 33.44 |
| LOOCV on signals from male subjects only | 1 | 66.56 |
| LOOCV on signals from dominant arm of female subjects only | 1 | 17.54 |
| LOOCV on signals from non-dominant arm of female subjects only | 1 | 17.67 |
| LOOCV on signals from dominant arm of male subjects only | 1 | 34.94 |
| LOOCV on signals from non-dominant arm of male subjects only | 1 | 31.62 |
| LOOCV on signals from a single subject only | 2 | 100 |
| LOOCV on signals from dominant arm of a single subject only | 2 | 54.89 |
| LOOCV on signals from non-dominant arm of a single subject only | 2 | 45.11 |
| Model | Data Type | Signals Truncated | Standard Deviation for All Data Types | Time for Training and Evaluation [h] | ||
|---|---|---|---|---|---|---|
| SVM | Hilbert transformed A-Scans | no | 85 | 1.95 | 0.17 | 88 |
| MLP | Hilbert transformed A-Scans | no | 84 | 1.97 | 6.66 | 88 |
| SVM | Fourier transformed A-Scans | no | 85 | 1.95 | 0.09 | 87 |
| MLP | Fourier transformed A-Scans | no | 84 | 1.97 | 6.12 | 87 |
| SVM | Wavelet transformed A-Scans | no | 85 | 1.95 | 0.12 | 86 |
| Evaluation Mode | ML Model | Data Type | Time for Evaluation and Training (Minutes) | |
|---|---|---|---|---|
| LOOCV | SVM | Wavelet transformed A-scans | 82 | 334 |
| LOOCV (dominant arm) | SVM | Wavelet transformed A-scans | 84 | 20 |
| LOOCV (non-dominant arm) | SVM | Combination of all possible features | 77 | <5 |
| LOOCV (female) | Logistic Regression | Combination of all possible features | 77 | <5 |
| LOOCV (female) [dominant arm] | Logistic Regression | Spectral features | 86 | <5 |
| LOOCV (female) [non-dominant arm] | Logistic Regression | Wavelet transformed A-Scans | 75 | <5 |
| LOOCV (male) | SVM | Wavelet transformed A-scans | 84 | 50 |
| LOOCV (male) [dominant arm] | SVM | Wavelet transformed A-scans | 86 | 5 |
| LOOCV (male) [non-dominant arm] | SVM | Combination of all possible features (of truncated signals) | 79 | <5 |
| LOOCV (single subject 09) | Logistic Regression | Statistical features | 70 | <5 |
| LOOCV (single subject 09) [dominant arm] | SVM | Wavelet transformed A-scans | 78 | 7 |
| LOOCV (single subject 09) [non-dominant arm] | SVM | Temporal features | 72 | <5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brausch, L.; Hewener, H.; Lukowicz, P. Classifying Muscle States with One-Dimensional Radio-Frequency Signals from Single Element Ultrasound Transducers. Sensors 2022, 22, 2789. https://doi.org/10.3390/s22072789
Brausch L, Hewener H, Lukowicz P. Classifying Muscle States with One-Dimensional Radio-Frequency Signals from Single Element Ultrasound Transducers. Sensors. 2022; 22(7):2789. https://doi.org/10.3390/s22072789
Chicago/Turabian StyleBrausch, Lukas, Holger Hewener, and Paul Lukowicz. 2022. "Classifying Muscle States with One-Dimensional Radio-Frequency Signals from Single Element Ultrasound Transducers" Sensors 22, no. 7: 2789. https://doi.org/10.3390/s22072789
APA StyleBrausch, L., Hewener, H., & Lukowicz, P. (2022). Classifying Muscle States with One-Dimensional Radio-Frequency Signals from Single Element Ultrasound Transducers. Sensors, 22(7), 2789. https://doi.org/10.3390/s22072789






