Collaborative Robot Precision Task in Medical Microbiology Laboratory
Abstract
:1. Introduction
2. Materials and Methods
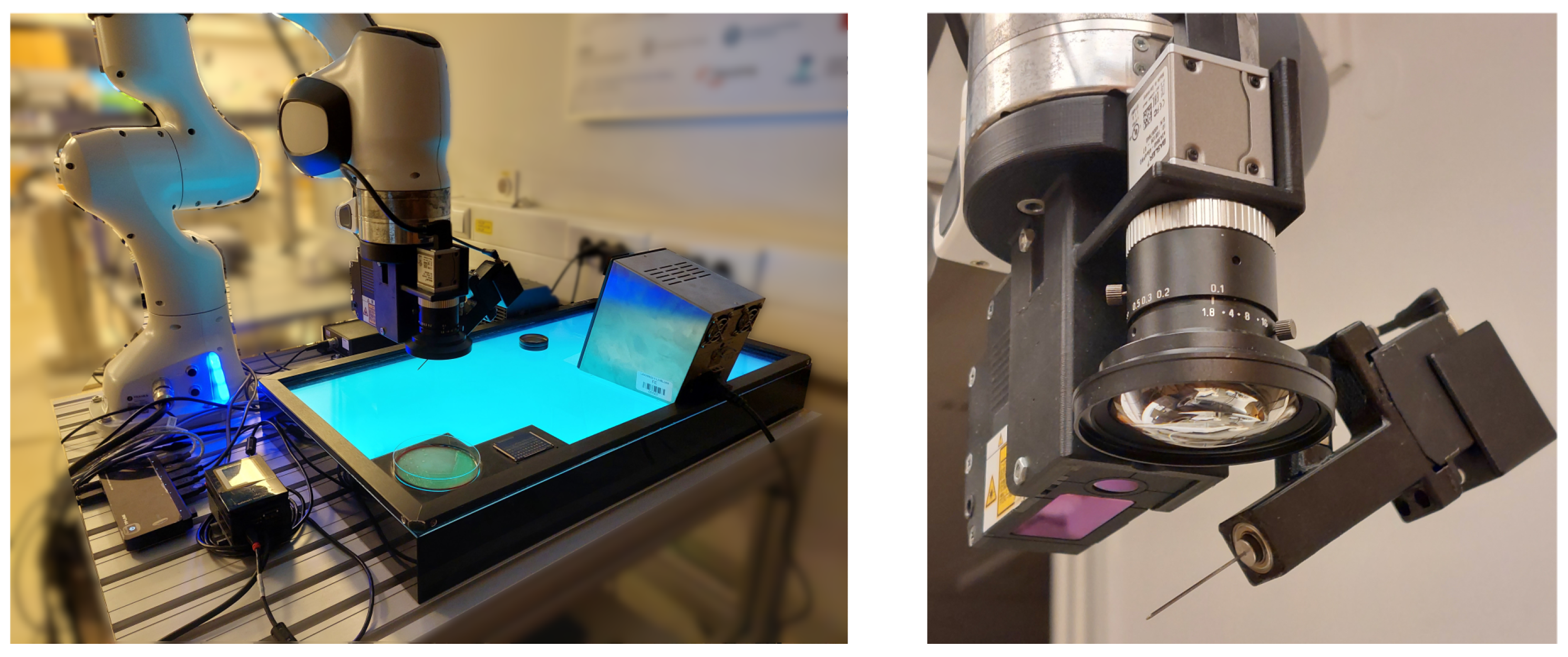
2.1. Experimental Setup
2.2. Learning from Demonstration and Trajectory Generation
2.3. Experiment Protocol
3. Results
3.1. Picking Process
3.2. Deposition Process
3.3. Identification
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| LfD | learning from demonstration |
| MS | mass spectrometry |
| MALDI | matrix-assisted laser desorption/ionization |
| TOF | time of flight |
| CRT | cooperative robot tool |
References
- Leber, A.L. Clinical Microbiology Procedures Handbook; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Buchan, B.W.; Ledeboer, N.A. Emerging technologies for the clinical microbiology laboratory. Clin. Microbiol. Rev. 2014, 27, 783–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, R. Matrix-assisted laser desorption ionization–time of flight mass spectrometry in clinical microbiology. Clin. Infect. Dis. 2013, 57, 564–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dadoun, R. Case study: Automation’s impact on productivity and turnaround time. MLO Med. Lab. Obs. 2002, 34, 36–38. [Google Scholar] [PubMed]
- Bizzini, A.; Durussel, C.; Bille, J.; Greub, G.; Prod’Hom, G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 2010, 48, 1549–1554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croxatto, A.; Prod’hom, G.; Faverjon, F.; Rochais, Y.; Greub, G. Laboratory automation in clinical bacteriology: What system to choose? Clin. Microbiol. Infect. 2016, 22, 217–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dauwalder, O.; Landrieve, L.; Laurent, F.; De Montclos, M.; Vandenesch, F.; Lina, G. Does bacteriology laboratory automation reduce time to results and increase quality management? Clin. Microbiol. Infect. 2016, 22, 236–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chudejova, K.; Bohac, M.; Skalova, A.; Rotova, V.; Papagiannitsis, C.C.; Hanzlickova, J.; Bergerova, T.; Hrabák, J. Validation of a novel automatic deposition of bacteria and yeasts on MALDI target for MALDI-TOF MS-based identification using MALDI Colonyst robot. PLoS ONE 2017, 12, e0190038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, P.; Watson, A.; Davies, M.; Stubbings, S. Integration of image analysis and robotics into a fully automated colony picking and plate handling system. Nucleic Acids Res. 1992, 20, 4599–4606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briner, D.R.; Sardhara, A.D.; Sugar, T.G. A Multi-Pin End-Effector for a Robotic Colony Picker. In Proceedings of the 2009 ASME Early Career Technical Conference, Tuscaloosa, AL, USA, 2–3 October 2009; pp. 222–228. [Google Scholar]
- Huang, C.; He, K.; Liu, C.; Fu, X.; Du, R. A colony picking robot with multi-pin synchronous manipulator. In Proceedings of the 2018 IEEE International Conference on Information and Automation (ICIA), Fujian, China, 11–13 August 2018; pp. 7–12. [Google Scholar]
- Fabritius, A.; Ng, D.; Kist, A.M.; Erdogan, M.; Portugues, R.; Griesbeck, O. Imaging-based screening platform assists protein engineering. Cell Chem. Biol. 2018, 25, 1554–1561. [Google Scholar] [CrossRef] [PubMed]
- Ravichandar, H.; Polydoros, A.S.; Chernova, S.; Billard, A. Recent advances in robot learning from demonstration. Annu. Rev. Control Robot. Auton. Syst. 2020, 3, 297–330. [Google Scholar] [CrossRef] [Green Version]
- Argall, B.D.; Chernova, S.; Veloso, M.; Browning, B. A survey of robot learning from demonstration. Robot. Auton. Syst. 2009, 57, 469–483. [Google Scholar] [CrossRef]
- Schou, C.; Damgaard, J.S.; Bøgh, S.; Madsen, O. Human-robot interface for instructing industrial tasks using kinesthetic teaching. In Proceedings of the IEEE ISR 2013, Seoul, Korea, 24–26 October 2013; pp. 1–6. [Google Scholar]
- Paraschos, A.; Daniel, C.; Peters, J.; Neumann, G. Using probabilistic movement primitives in robotics. Auton. Robot. 2018, 42, 529–551. [Google Scholar] [CrossRef]
- Steinmetz, F.; Montebelli, A.; Kyrki, V. Simultaneous kinesthetic teaching of positional and force requirements for sequential in-contact tasks. In Proceedings of the 2015 IEEE-RAS 15th International Conference on Humanoid Robots (Humanoids), Seoul, Korea, 3–5 November 2015; pp. 202–209. [Google Scholar]
- Berio, D.; Calinon, S.; Leymarie, F.F. Learning dynamic graffiti strokes with a compliant robot. In Proceedings of the 2016 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), Daejeon, Korea, 9–14 October 2016; pp. 3981–3986. [Google Scholar]
- Kronander, K.; Billard, A. Learning compliant manipulation through kinesthetic and tactile human-robot interaction. IEEE Trans. Haptics 2013, 7, 367–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calinon, S.; Bruno, D.; Malekzadeh, M.S.; Nanayakkara, T.; Caldwell, D.G. Human—Robot skills transfer interfaces for a flexible surgical robot. Comput. Methods Programs Biomed. 2014, 116, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Ye, M.; Hu, Y.; Vandini, A.; Lee, S.L.; Yang, G.Z. A multirobot cooperation framework for sewing personalized stent grafts. IEEE Trans. Ind. Inform. 2017, 14, 1776–1785. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; Mahler, J.; Laskey, M.; Li, P.; Goldberg, K. Using dVRK teleoperation to facilitate deep learning of automation tasks for an industrial robot. In Proceedings of the 2017 13th IEEE Conference on Automation Science and Engineering (CASE), Xian, China, 20–23 August 2017; pp. 1–8. [Google Scholar]
- Abbott, J.J.; Hager, G.D.; Okamura, A.M. Steady-hand teleoperation with virtual fixtures. In Proceedings of the 12th IEEE International Workshop on Robot and Human Interactive Communication, ROMAN, Millbrae, CA, USA, 31 October–2 November 2003; pp. 145–151. [Google Scholar]
- Bettini, A.; Marayong, P.; Lang, S.; Okamura, A.M.; Hager, G.D. Vision-assisted control for manipulation using virtual fixtures. IEEE Trans. Robot. 2004, 20, 953–966. [Google Scholar] [CrossRef] [Green Version]
- Baumkircher, A.; Munih, M.; Mihelj, M. Performance analysis of learning from demonstration approaches during a fine movement generation. IEEE Trans. Hum.-Mach. Syst. 2021, 51, 653–662. [Google Scholar] [CrossRef]
- Ijspeert, A.J.; Nakanishi, J.; Hoffmann, H.; Pastor, P.; Schaal, S. Dynamical movement primitives: Learning attractor models for motor behaviors. Neural Comput. 2013, 25, 328–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calinon, S.; Guenter, F.; Billard, A. On learning, representing, and generalizing a task in a humanoid robot. IEEE Trans. Syst. Man, Cybern. Part B (Cybern.) 2007, 37, 286–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Rozo, L.; Silvério, J.; Caldwell, D.G. Kernelized movement primitives. Int. J. Robot. Res. 2019, 38, 833–852. [Google Scholar] [CrossRef] [Green Version]
- Paraschos, A.; Daniel, C.; Peters, J.; Neumann, G. Probabilistic movement primitives. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2013. [Google Scholar]




| ID | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Initial volume [mm] | 1.44 | 0.78 | 0.46 | 0.11 | 0.06 | 0.02 |
| Final volume [mm] | 0.41 | −0.04 | 0.04 | 0.05 | 0.007 | −0.02 |
| Relative change [%] | −72 | −105 | −91 | −54 | −99 | −193 |
| Acinetobacter baumannii | Staphylococcus epidermidis | |
|---|---|---|
| No. of samples | 36 | 20 |
| No. of “score below 1.70” | 3 | 1 |
| No. of “no peaks found” | 2 | 0 |
| No. of total errors | 5 | 1 |
| Total errors [%] | 13.9 | 5 |
| Average identification score w/o errors | 2.05 ± 0.21 | 2.06 ± 0.13 |
| Average identification score with errors | 1.94 ± 0.54 | 2.04 ± 0.16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baumkircher, A.; Seme, K.; Munih, M.; Mihelj, M. Collaborative Robot Precision Task in Medical Microbiology Laboratory. Sensors 2022, 22, 2862. https://doi.org/10.3390/s22082862
Baumkircher A, Seme K, Munih M, Mihelj M. Collaborative Robot Precision Task in Medical Microbiology Laboratory. Sensors. 2022; 22(8):2862. https://doi.org/10.3390/s22082862
Chicago/Turabian StyleBaumkircher, Aljaz, Katja Seme, Marko Munih, and Matjaž Mihelj. 2022. "Collaborative Robot Precision Task in Medical Microbiology Laboratory" Sensors 22, no. 8: 2862. https://doi.org/10.3390/s22082862






