Harmful Microalgae Detection: Biosensors versus Some Conventional Methods
Abstract
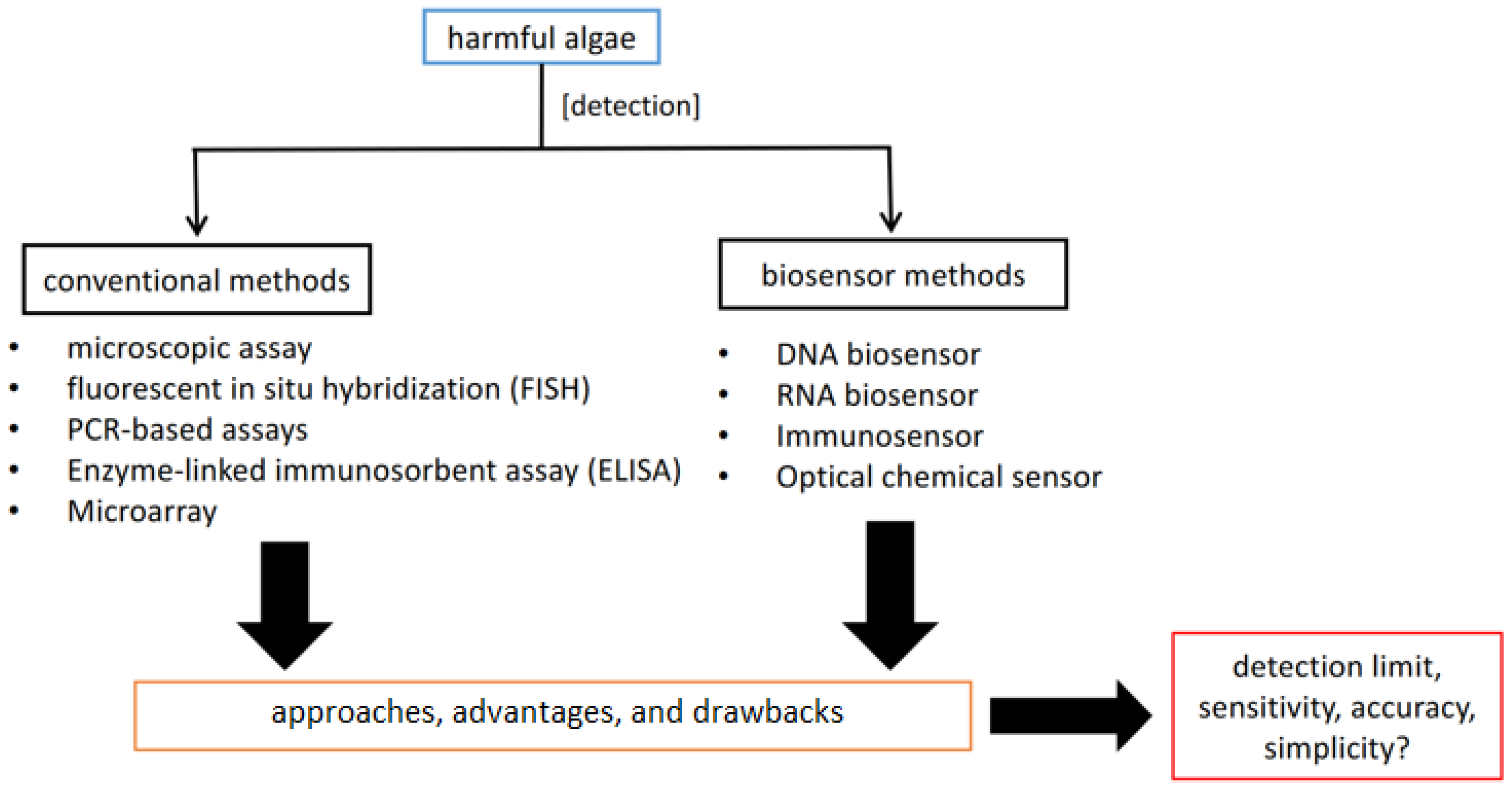
1. Introduction
2. Conventional Methods for HAB Identification
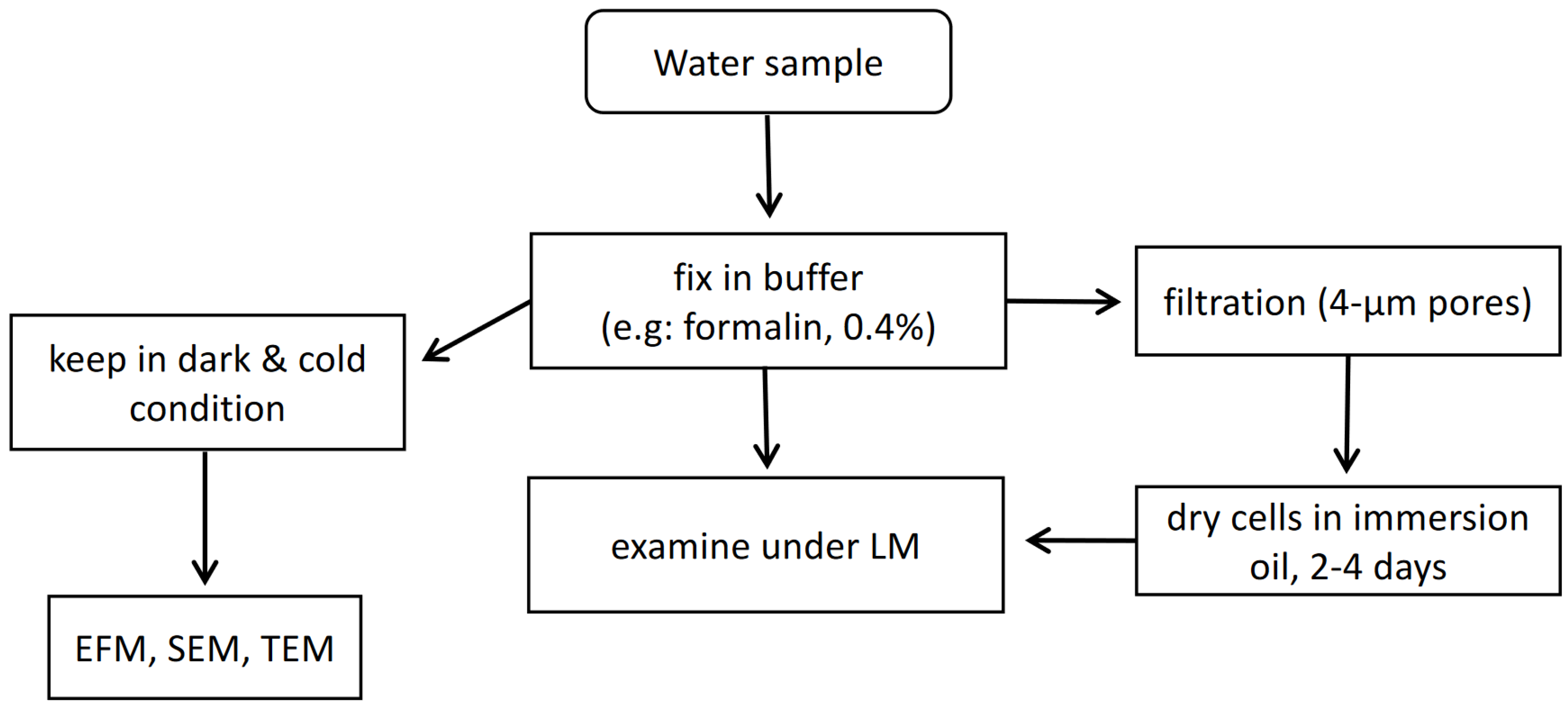
2.1. Microscopic Analysis
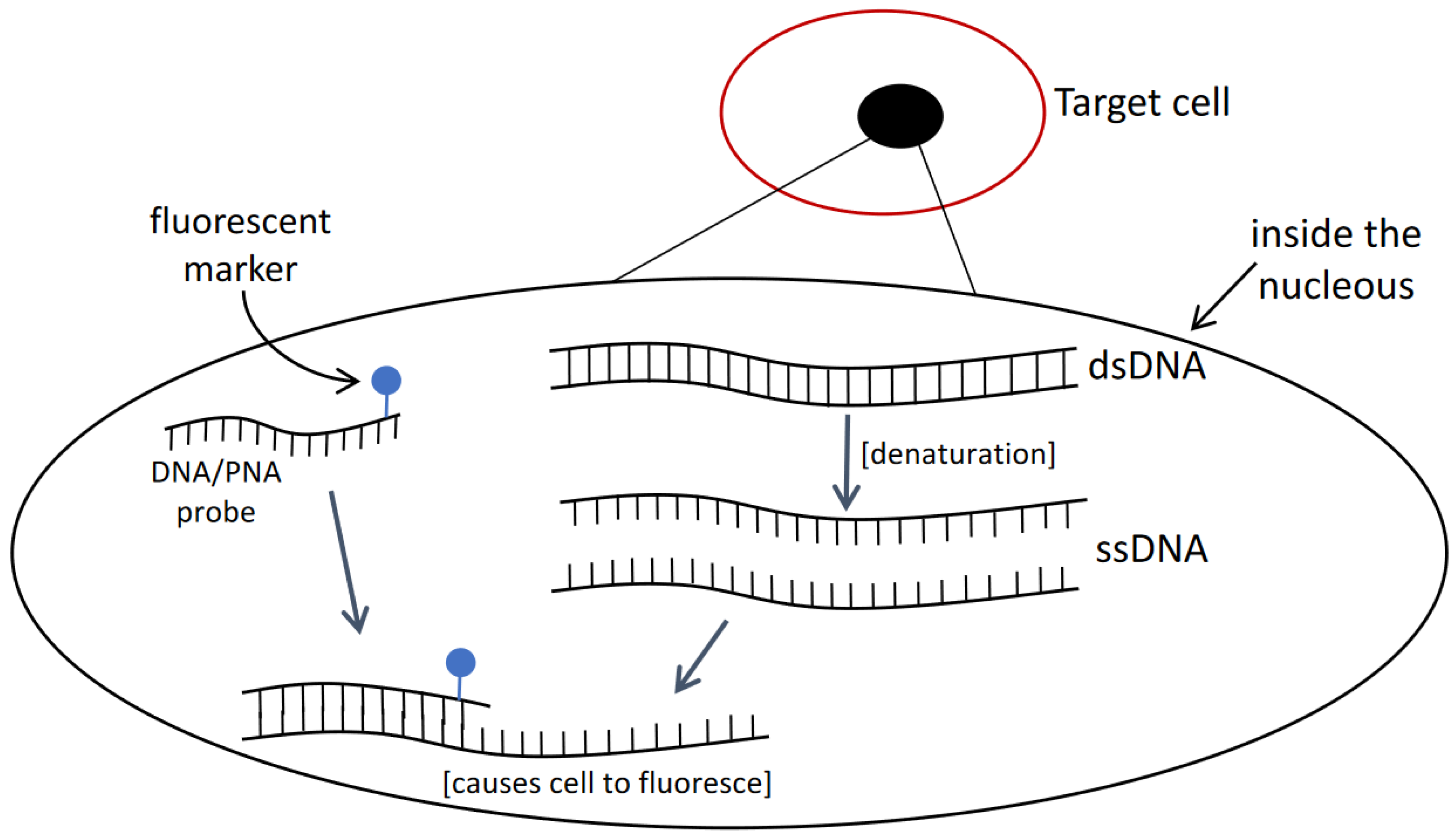
2.2. Fluorescent In Situ Hybridization (FISH)
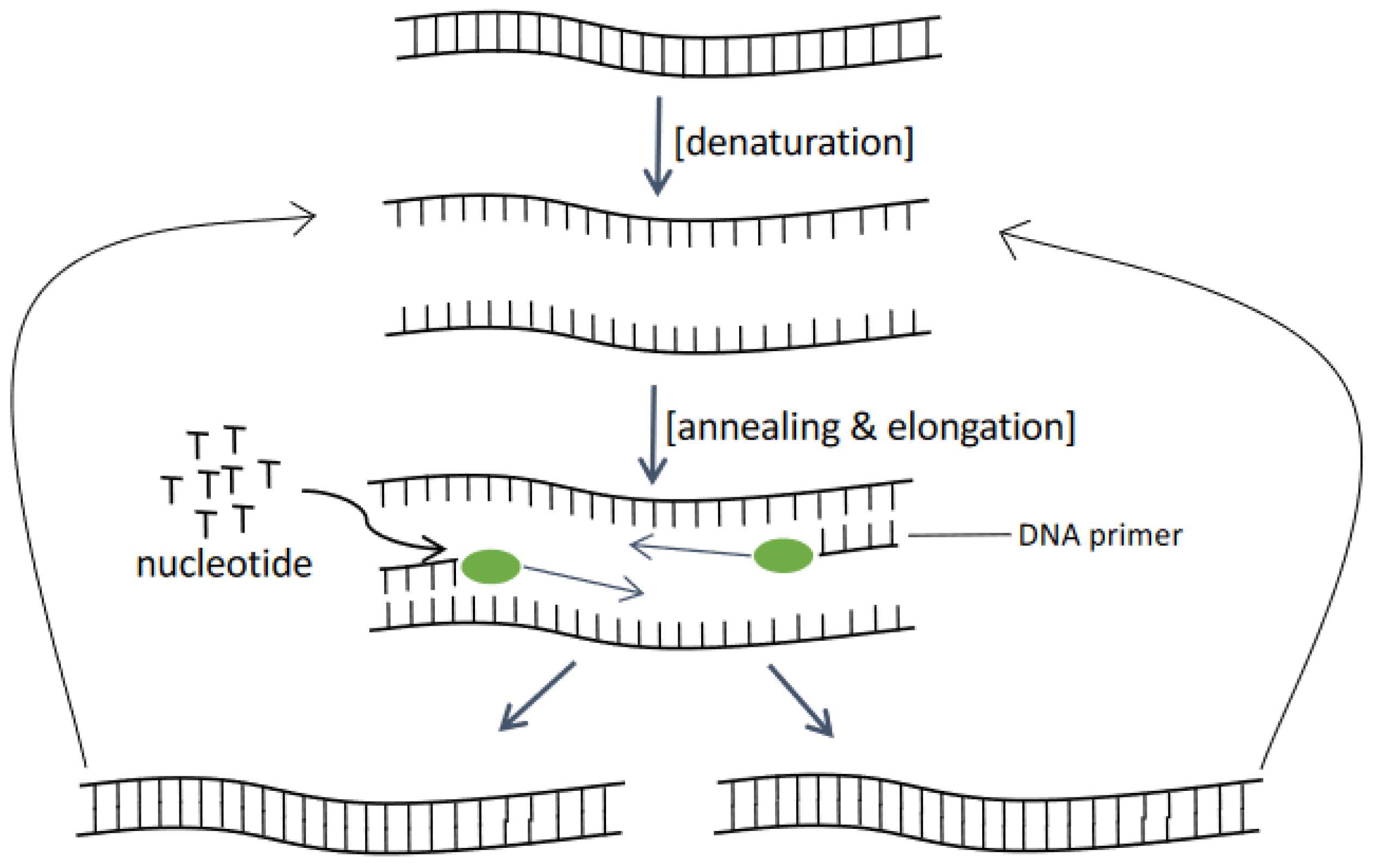
2.3. Polymerase Chain Reaction (PCR)-Based Assays
2.4. Enzyme-Linked Immunosorbent Assay (ELISA)
2.5. Microarray
3. Biosensor Methods
3.1. Electrochemical Biosensor Methods
3.2. Optical Biosensors
4. Nanomaterials in Biosensor
4.1. Nanomaterial-Based Immunosensor
4.2. Nanomaterial-Based DNA Biosensor
5. A Detailed Comparison of Biosensor and Conventional Methods for Harmful Microalgae Determination
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Laabir, M.; Jauzein, C.; Genovesi, B.; Masseret, E.; Cecchi, P.; Collos, Y. Influence of temperature, salinity and irradiance on the growth and cell yield of the harmful red tide dinoflagellate Alexandrium catenella colonizing Mediterranean waters. J. Plankton. Res. 2011, 33, 1550–1563. [Google Scholar] [CrossRef]
- Adam, A.; Mohammad-Noor, N.; Anton, A.; Saleh, E.; Saad, S.; Muhd Shaleh, S.R. Temporal and spatial distribution of harmful algal bloom (HAB) species in coastal waters of Kota Kinabalu, Sabah, Malaysia. Harmful Algae 2011, 10, 495–502. [Google Scholar] [CrossRef]
- Dale, B.; Edwards, M.; Reid, P.C. Climate Change and Harmful Algal Blooms. In Ecology of Harmful Algae; Springer: Berlin/Heidelberg, Germany, 2006; pp. 367–378. [Google Scholar]
- Handy, S.M.; Demir, E.; Hutchins, D.A.; Portune, K.J.; Whereat, E.B.; Hare, C.E.; Rose, J.M.; Warner, M.; Farestad, M.; Cary, S.C. Using quantitative real-time PCR to study competition and community dynamics among Delaware Inland Bays harmful algae in field and laboratory studies. Harmful Algae 2008, 7, 599–613. [Google Scholar] [CrossRef]
- Bricker, S.B.; Longstaff, B.; Dennison, W.; Jones, A.; Boicourt, K.; Wicks, C.; Woerner, J. Effects of nutrient enrichment in the nation’s estuaries: A decade of change. Harmful Algae 2008, 8, 21–32. [Google Scholar] [CrossRef]
- Paerl, H.W.; Gardner, W.S.; Havens, K.E.; Joyner, A.R.; Mccarthy, M.J.; Newell, S.E.; Qin, B.; Scott, J.T. Mitigating cyanobacterial harmful algal blooms in aquatic ecosystems impacted by climate change and anthropogenic nutrients. Harmful Algae 2016, 54, 213–222. [Google Scholar] [CrossRef]
- O’neil, J.; Davis, T.W.; Burford, M.A.; Gobler, C. The rise of harmful cyanobacteria blooms: The potential roles of eutrophication and climate change. Harmful Algae 2012, 14, 313–334. [Google Scholar] [CrossRef]
- Branch, G.M.; Bustamante, R.H.; Robinson, T.B. Impacts of a ‘black tide’ harmful algal bloom on rocky-shore intertidal communities on the West Coast of South Africa. Harmful Algae 2013, 24, 54–64. [Google Scholar] [CrossRef]
- Zhen, Y.; Mi, T.; Yu, Z. Detection of several harmful algal species by sandwich hybridization integrated with a nuclease protection assay. Harmful Algae 2009, 8, 651–657. [Google Scholar] [CrossRef]
- Meneely, J.P.; Campbell, K.; Greef, C.; Lochhead, M.J.; Elliott, C.T. Development and validation of an ultrasensitive fluorescence planar waveguide biosensor for the detection of paralytic shellfish toxins in marine algae. Biosens. Bioelectron. 2013, 41, 691–697. [Google Scholar] [CrossRef][Green Version]
- Medlin, L. Molecular tools for monitoring harmful algal blooms. Environ. Sci. Pollut. Res. 2013, 20, 6683–6685. [Google Scholar] [CrossRef]
- Gas, F.; Baus, B.; Pinto, L.; Compere, C.; TanchoU, V.; Quéméneur, E. One step immunochromatographic assay for the rapid detection of Alexandrium minutum. Biosens. Bioelectron. 2010, 25, 1235–1239. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dittami, S.M.; Hostyeva, V.; Egge, E.S.; Kegel, J.U.; Eikrem, W.; Edvardsen, B. Seasonal dynamics of harmful algae in outer Oslofjorden monitored by microarray, qPCR, and microscopy. Environ. Sci. Pollut. R. 2013, 20, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Orozco, J.; Medlin, L.K. Review: Advances in electrochemical genosensors-based methods for monitoring blooms of toxic algae. Environ. Sci. Pollut. R. 2013, 20, 6838–6850. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.T. Planktonic marine copepods and harmful algae. Harmful Algae 2014, 32, 81–93. [Google Scholar] [CrossRef]
- Berdalet, E.; Fleming, L.E.; Gowen, R.; Davidson, K.; Hess, P.; Backer, L.C.; Moore, S.K.; Hoagland, P.; Enevoldsen, H. Marine harmful algal blooms, human health and wellbeing: Challenges and opportunities in the 21st century. J. Mar. Biol. Assoc. UK 2015, 96, 61–91. [Google Scholar] [CrossRef] [PubMed]
- Hallegraeff, G.M. A review of harmful algal blooms and their apparent global increase. Phycologia 1993, 32, 79–99. [Google Scholar] [CrossRef]
- Ajani, P.; Harwoood, D.T.; Murray, S.A. Recent trends in marine phycotoxins from Australian coastal waters. Mar. Drugs 2017, 15, 33. [Google Scholar] [CrossRef]
- Lucas, B.; Dahlmann, J.; Erler, K.; Gerdts, G.; Wasmund, N.; Hummert, C. Overview of key phytoplankton toxins and their recent occurrence in the North and Baltic Sea. Environ. Toxicol. 2005, 20, 1–17. [Google Scholar] [CrossRef]
- Watkins, S.M.; Reich, A.; Fleming, L.E.; Hammond, R. Neurotoxic Shellfish Poisoning. Mar. Drugs 2008, 6, 431–455. [Google Scholar] [CrossRef]
- Kouakou, C.R.C.; Poder, T.G. Economic impact of harmful algal blooms on human health: A systematic review. J. Water Health 2019, 17, 499–516. [Google Scholar] [CrossRef]
- Teen, L.P.; Gires, U.; Pin, L.C. Harmful Algal Blooms in Malaysian Waters. Sains Malays. 2012, 41, 1509–1515. [Google Scholar]
- Diercks, S.; Metfies, K.; Medlin, L.K. Development and adaptation of a multiprobe biosensor for the use in a semi-automated device for the detection of toxic algae. Biosens. Bioelectron. 2008, 23, 1527–1533. [Google Scholar] [CrossRef]
- Cox, A.M.; Goodwin, K.D. Sample preparation methods for quantitative detection of DNA by molecular assays and marine biosensors. Mar. Pollut. Bull. 2013, 73, 47–56. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, C.; Wang, Y.; Chen, G. A review of the current and emerging detection methods of marine harmful microalgae. Sci. Total Environ. 2022, 815, 152913. [Google Scholar] [CrossRef] [PubMed]
- Jipanin, S.J.; Shaleh, S.R.M.; Lim, P.T.; Leaw, C.P.; Mustapha, S. The Monitoring of Harmful Algae Blooms in Sabah, Malaysia. J. Phys. Conf. Ser. 2019, 1358, 1–9. [Google Scholar] [CrossRef]
- Scorzetti, G.; Brand, L.; Hitchcock, G.; Rein, K.; Sinigalliano, C.; Fell, J. Multiple simultaneous detection of Harmful Algal Blooms (HABs) through a high throughput bead array technology, with potential use in phytoplankton community analysis. Harmful Algae 2009, 8, 196–211. [Google Scholar] [CrossRef]
- Antonella, P.; Luca, G. The quantitative real-time PCR applications in the monitoring of marine harmful algal bloom (HAB) species. Environ. Sci. Pollut. R. 2012, 20, 1–12. [Google Scholar] [CrossRef]
- Scholin, C. The development and application of molecular probes and novel instrumentation for detection of harmful algae. In Proceedings of the Ocean Development & International Law, Nice, France, 1 October 1998; pp. 367–370. [Google Scholar]
- Scholin, C.; Doucette, G.; Cembella, A. Prospects for developing automated systems for in situ detection of harmful algae and their toxins. In Real-time Coastal Observing Systems for Marine Ecosystem Dynamics and Harmful Algal Blooms; The United Nations Educational, Scientific and Cultural Organization: Paris, France, 2008; pp. 413–462. [Google Scholar]
- Metfies, K.; Töbe, K.; Scholin, C.; Medlin, L.K. Laboratory and Field Applications of Ribosomal RNA Probes to Aid the Detection and Monitoring of Harmful Algae. Ecol. Harmful Algae 2006, 189, 311–325. [Google Scholar]
- Barlaan, E.A.; Furukawa, S.; Takeuchi, K. Detection of bacteria associated with harmful algal blooms from coastal and microcosm environments using electronic microarrays. Environ. Microbiol. 2007, 9, 690–702. [Google Scholar] [CrossRef]
- Diercks, S.; Gescher, C.; Metfies, K.; Medlin, L.K. Evaluation of locked nucleic acids for signal enhancement of oligonucleotide probes for microalgae immobilised on solid surfaces. J. Appl. Phycol. 2009, 21, 657–668. [Google Scholar] [CrossRef]
- Orozco, J.; Medlin, L.K. Electrochemical performance of a DNA-based sensor device for detecting toxic algae. Sens. Actuators B-Chem. 2011, 153, 71–77. [Google Scholar] [CrossRef]
- Diercks-Horn, S.; Metfies, K.; Jäckel, S.; Medlin, L.K. The ALGADEC device: A semi-automated rRNA biosensor for the detection of toxic algae. Harmful Algae 2011, 10, 395–401. [Google Scholar] [CrossRef]
- Booth, B.C. Estimating Cell Concentration and Biomass of Autotrophic Plankton Using Microscopy. In Handbook of Methods in Aquatic Microbial Ecology; CRC Press: Boca Raton, FL, USA, 2018; pp. 199–205. [Google Scholar]
- Usup, G.; Pin, L.C.; Ahmad, A.; Teen, L.P. Alexandrium (Dinophyceae) species in Malaysian waters. Harmful Algae 2002, 1, 265–275. [Google Scholar] [CrossRef]
- Lorenzen, C.J. A method for the continuous measurement of in vivo chlorophyll concentration. Deep-Sea Res. 1966, 13, 223. [Google Scholar] [CrossRef]
- Legner, M. Phytoplankton quantity assessment by means of flow cytometry. Mar. Microb. Food Webs 1990, 4, 161. [Google Scholar]
- Sako, Y.; Hosoi-Tanabe, S.; Uchida, A. Fluorescence in situ hybridization using rrna-targeted probes for simple and rapid identification of the toxic dinoflagellates Alexandrium tamarense and Alexandrium catenella. J. Phycol. 2004, 40, 598–605. [Google Scholar] [CrossRef]
- Miller, P.E.; Scholin, C.A. Identification and enumeration of cultured and wild Pseudo-nitzschia probes and filter-based whole cell hybridization. J. Phycol. 1998, 34, 371–382. [Google Scholar] [CrossRef]
- Medlin, L.K.; Orozco, J. Molecular techniques for the detection of organisms in aquatic environments, with emphasis on harmful algal bloom species. Sensors 2017, 17, 1184. [Google Scholar] [CrossRef]
- Yek, L.H.; Hii, K.S.; Tan, T.H.; Kon, N.F.; Lim, P.T.; Leaw, C.P. Rapid detection of toxic dinoflagellate, Alexandrium minutum (Dinophyceae) using fluorescence in situ hybridization (FISH). Malays. J. Sci. 2013, 32, 1–12. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, C.; Zhang, B.; Wang, G.; Lu, D.; Xu, Z.; Yan, P. Development of a PNA probe for fluorescence in situ hybridization detection of Prorocentrum donghaiense. PLoS ONE 2011, 6, e25527. [Google Scholar] [CrossRef]
- Sun, Y.J.; Chen, G.F.; Zhang, C.Y.; Guo, C.L.; Wang, Y.Y.; Sun, R. Development of a multiplex polymerase chain reaction assay for the parallel detection of harmful algal bloom-forming species distributed along the Chinese coast. Harmful Algae 2019, 84, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Pearson, L.A.; D’Agostino, P.M.; Neilan, B.A. Recent developments in quantitative PCR for monitoring harmful marine microalgae. Harmful Algae 2021, 108, 102096. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, C.T.; Herrmann, M.G.; Moss, A.A.; Rasmussen, R.P. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 1997, 22, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yu, R.C.; Chen, J.H.; Zhang, Q.C.; Kong, F.Z.; Zhou, M.J. Distribution of Alexandrium fundyense and A.pacificum (Dinophycease) in the Yellow Sea and Bohai Sea. Mar. Pollut. Bull. 2015, 96, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Gas, F.; Pinto, L.; Baus, B.; Gaufres, L.; Crassous, M.P.; Compere, C.; Quéméneur, E. Monoclonal antibody against the surface of Alexandrium minutum used in a whole-cell ELISA. Harmful Algae 2009, 8, 538–545. [Google Scholar] [CrossRef]
- Anderson, D.M.; Kulis, D.; Erdner, D.; Ahn, S.; Walt, D. Fibre optic microarrays for the detection and enumeration of harmful algal bloom species. Afr. J. Mar. Sci. 2006, 28, 231–235. [Google Scholar] [CrossRef]
- Ahn, S.; Kulis, D.M.; Erdner, D.L.; Anderson, D.M.; Walt, D.R. Fiber-Optic Microarray for Simultaneous Detection of Multiple Harmful Algal Bloom Species. Appl. Environ. Microbiol. 2006, 72, 5742–5749. [Google Scholar] [CrossRef]
- Walt, D.R. Bead-based fiber optic arrays. Science 2000, 287, 451–452. [Google Scholar] [CrossRef]
- Naresh, V.; Lee, N. Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef]
- Andryukov, B.G.; Besednova, N.N.; Romashko, R.V.; Zaporozhets, T.S.; Efimov, T.A. Label-Free Biosensors for Laboratory-Based Diagnostics of Infections: Current Achievements and New Trends. Biosensors 2020, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Pilas, J.; Yazici, Y.; Selmer, T.; Keusgen, M.; Schöning, M.J. Application of a Portable Multi-Analyte Biosensor for Organic Acid Determination in Silage. Sensors 2018, 18, 1470. [Google Scholar] [CrossRef] [PubMed]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar] [PubMed]
- Wu, Q.; Zhang, Y.; Yang, Q.; Yuan, N.; Zhang, W. Review of Electrochemical DNA Biosensors for Detecting Food Borne Pathogens. Sensors 2019, 19, 4916. [Google Scholar] [CrossRef]
- Hosu, O.; Selvolini, G.; Cristea, C.; Marrazza, G. Electrochemical Immunosensors for Disease Detection and Diagnosis. Curr. Med. Chem. 2018, 25, 4119–4137. [Google Scholar] [CrossRef]
- Sokolov, A.N.; Roberts, M.E.; Bao, Z. Fabrication Of Low-Cost Electronic Biosensors. Mater. Today 2009, 12, 12–20. [Google Scholar] [CrossRef]
- da Silva, T.S.G.; Souto, E.P.; Barragan, T.C.; Giarola, D.F.; de Moraes, A.C.M.; Kubota, L.T. Electrochemical Biosensors in Point-of-Care Devices: Recent Advances and Future Trends. Chemelectrochem 2017, 4, 778–794. [Google Scholar] [CrossRef]
- Wojciechowski, M.F.; Sanderson, M.J.; Hu, J.M. Evidence on the monophyly of Astragalus (Fabaceae) and its major subgroups based on nuclear ribosomal DNA ITS and chloroplast DNA trnL intron data. Syst. Bot. 1999, 24, 409–437. [Google Scholar] [CrossRef]
- Lagier, M.J.; Fell, J.W.; Goodwin, K.D. Electrochemical detection of harmful algae and other microbial contaminants in coastal waters using hand-held biosensors. Mar. Pollut. Bull. 2007, 54, 757–770. [Google Scholar] [CrossRef]
- Metfies, K.; Huljic, S.; Lange, M.; Medlin, L.K. Electrochemical detection of the toxic dinoflagellate Alexandrium ostenfeldii with a DNA-biosensor. Biosens. Bioelectron. 2005, 20, 1349–1357. [Google Scholar] [CrossRef]
- Orozco, J.; Baudart, J.; Medlin, L.K. Evaluation of probe orientation and effect of the digoxigenin-enzymatic label in a sandwich hybridization format to develop toxic algae biosensors. Harmful Algae 2011, 10, 489–494. [Google Scholar] [CrossRef]
- Bratcher, A.R.; Connell, L.B. The Use of Peptide Nucleic Acids in Surface Plasmon Resonance for Detection of Red Tide Algae. In Molecular Biological Technologies for Ocean Sensing; Humana Press: Totova, NJ, USA, 2012. [Google Scholar]
- Lucarelli, F.; Tombelli, S.; Minunni, M.; Marrazza, G.; Mascini, M. Electrochemical and piezoelectric DNA biosensors for hybridisation detection. Anal. Chim. Acta 2008, 609, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Mannelli, I.; Lecerf, L.; Guerrouache, M.; Goossens, M.; Millot, M.C.; Canva, M. DNA immobilisation procedures for surface plasmon resonance imaging (SPRI) based microarray systems. Biosens. Bioelectron. 2007, 22, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Mastali, M.; Li, Y.; Gau, V. Development of an Advanced Electrochemical DNA Biosensor for Bacterial Pathogen Detection. J. Mol. Diagn. 2007, 9, 158–168. [Google Scholar] [CrossRef]
- Fowler, J.M.; Stuart, M.C.; Wong, D.K. Self-assembled layer of thiolated protein G as an immunosensor scaffold. Anal. Chem. 2007, 79, 350–354. [Google Scholar] [CrossRef]
- Aydin, M.; Aydin, E.B.; Sezginturk, M.K. Advances in immunosensor technology. Adv. Clin. Chem. 2020, 102, 1–62. [Google Scholar]
- Oloketuyi, S.; Mazzega, E.; Zavasnik, J.; Pungjunun, K.; Kalcher, K.; Marco, A.D.; Mehmeti, E. Electrochemical immunosensor functionalized with nanobodies for the detection of the toxic microalgae Alexandrium minutum using glassy carbon electrode modified with gold nanoparticles. Biosens. Bioelectron. 2020, 154, 112052. [Google Scholar] [CrossRef]
- Morgan, C.L.; Newman, D.J.; Price, C. Immunosensors: Technology and opportunities in laboratory medicine. Clin. Chem. 1996, 42, 193–209. [Google Scholar] [CrossRef]
- Stauffer, B.A.; Bowers, H.; Buckley, E.; Davis, T.W. Considerations in Harmful Algal Bloom Research and Monitoring: Perspectives From a Consensus-Building Workshop and Technology Testing. Front. Mar. Sci. 2019, 6, 1–12. [Google Scholar] [CrossRef]
- Lei, Y.; Chen, W.; Mulchandani, A. Microbial biosensors. Anal. Chim. Acta 2006, 568, 200–210. [Google Scholar] [CrossRef]
- Leiner, M.J.; Hartmann, P. Theory and practice in optical pH sensing. Sens. Actuators B-Chem. 1993, 11, 281–289. [Google Scholar] [CrossRef]
- Auffan, M.; Rose, J.; Bottero, J.Y.; Lowry, G.V.; Jolivet, J.P.; Wiesner, M.R. Towards a definition of inorganic nanoparticles from an environmental, health and safety perspective. Nat. Nanotechnol. 2009, 4, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Cheon, J.; Lee, J.H. Synergistically integrated nanoparticles as multimodal probes for nanobiotechnology. Accounts Chem. Res. 2008, 41, 1630–1640. [Google Scholar] [CrossRef] [PubMed]
- Boonkaew, S.; Chaiyo, S.; Jampasa, S.; Rengpipat, S.; Siangproh, W.; Chailapakul, O. An origami paper-based electrochemical immunoassay for the C-reactive protein using a screen-printed carbon electrode modifed with graphene and gold nanoparticles. Microchim. Acta. 2019, 186, 153. [Google Scholar] [CrossRef] [PubMed]
- Mazzega, E.; Beran, A.; Cabrini, M.; Marco, A.D. In vitro isolation of nanobodies for selective Alexandrium minutum recognition: A mondel for convenient development of dedicated immuno-reagents to study and diagnostic toxic unicellular algae. Harmful Algae 2019, 82, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, 690–697. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.P.; Liu, J.L.; Shriver-Lake, L.C.; Zabetakis, D.; Sugiharto, V.A.; Chen, H.W.; Lee, C.R.; Defang, G.N.; Wu, S.J.L.; Venkateswaran, N.; et al. Oriented Immobilization of Single-Domain Antibodies Using SpyTag/SpyCatcher Yields Improved Limits of Detection. Anal. Chem. 2019, 91, 9424–9429. [Google Scholar] [CrossRef]
- Heukers, R.; Altintas, I.; Raghoenath, S.; Zan, E.D.; Pepermans, R.; Roovers, R.C.; Haselberg, R.; Hennink, W.E.; Schiffelers, R.M.; Kok, R.J.; et al. Targeting hepatocyte growth factor receptor (Met) positive tumor cells using internalizing nanobody-decorated albumin nanoparticles. Biomaterials 2014, 35, 601–610. [Google Scholar] [CrossRef]
- Crépin, R.; Gentien, D.; Duché, A.; Rapinat, A.; Reyes, C.; Némati, F.; Massonnet, G.; Decaudin, D.; Djender, S.; Moutel, S.; et al. Nanobodies against surface biomarkers enable the analysis of tumor genetic heterogeneity in uveal melanoma patient-derived xenografts. Pigment. Cell Melanoma Res. 2017, 30, 3017–3327. [Google Scholar] [CrossRef]
- Niemeyer, C.M.; Simon, U. DNA-Based Assembly of Metal Nanoparticles. Eur. J. Inorg. Chem. 2005, 2005, 3641–3655. [Google Scholar] [CrossRef]
- Salam, F.; Razali, H.; Rahman, G.A.; Razali, R.; Ayub, M.N.A.; Ramli, S.N.M. Development of Immunosensor for Early Detection of Aquatic Harmful Algal Blooms (HABs). Mater. Sci. Eng. 2017, 7, 127–134. [Google Scholar]
- Jacobs, M.; Selvam, A.P.; Craven, J.E.; Prasad, S. Antibody-Conjugated Gold Nanoparticle-Based Immunosensor for Ultra-Sensitive Detection of Troponin-T. J. Lab. Autom. 2014, 19, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Lepoitevin, M.; Lemouel, M.; Bechelany, M.; Janot, J.M.; Balme, S. Gold nanoparticles for the bare-eye based and spectrophotometric detection of proteins, polynucleotides and DNA. Microchim. Acta 2015, 182, 1223–1229. [Google Scholar] [CrossRef]
- Jang, H.; Kwak, C.H.; Kim, S.M.; Huh, Y.S.; Jeon, T.J. Identification of genetically modified DNA found in Roundup Ready soybean using gold nanoparticles. Microchim. Acta. 2016, 183, 2649–2654. [Google Scholar] [CrossRef]
- Zhang, F.; Shi, Y.; Jiang, K.; Xu, Z.; Ma, L. Sensitive and rapid detection of two toxic microalgae Alexandrium by loop-mediated isothermal amplifification. Acta. Oceanol. Sin. 2012, 31, 139–146. [Google Scholar] [CrossRef]
- Sousa, C.; Compère, C.; Dreanno, C.; Crassous, M.; Gas, F.; Baus, B.; Perrot, H. Direct and fast detection of Alexandrium minutum algae by using high frequency microbalance. J. Microbiol. Meth. 2014, 104, 49–54. [Google Scholar] [CrossRef]
- Gas, F.; Baus, B.; Queré, J.; Chapelle, A.; Dreanno, C. Rapid detection and quantifification of the marine toxic algae, Alexandrium minutum, using a super-paramagnetic immunochromatographic strip test. Talanta 2016, 147, 581–589. [Google Scholar] [CrossRef]
- Wang, J. Electrochemical biosensing based on noble metal nanoparticles. Microchim. Acta 2012, 177, 245–270. [Google Scholar] [CrossRef]
- Das, S.; Mitra, S.; Khurana, S.M.P.; Debnath, N. Nanomaterials for biomedical applications. Front Life Sci. 2013, 7, 90–98. [Google Scholar] [CrossRef]
- Sharma, R.; Ragavan, K.; Thakur, M.; Raghavarao, K. Recent advances in nanoparticle based aptasensors for food contaminants. Biosens. Bioelectron. 2015, 74, 612–627. [Google Scholar] [CrossRef]
- Qiang, L.; Zhang, Y.; Wu, C.; Han, Y.; Wang, S.; Wang, Y.; Zhang, C.; Liu, G.; Wu, Q.; Liu, H.; et al. A Facile and Sensitive DNA Sensing of Harmful Algal Blooms Based on Graphene Oxide Nanosheets. Mar. Biotechnol. 2020, 22, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, M.; Xu, X.; Zhang, L. Graphene oxide reduced and modified by environmentally friendly glyclyglycine and its excellent catalytic performance. Nanotechnology 2014, 25, 135707. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Qiu, S.; Du, P.; Zhao, H.; Wang, L. An ionic liquid-graphene oxide hybrid nanomaterial: Synthesis and anticorrosive applications. Nanoscale 2018, 10, 8115–8124. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Meng, G.X.; Zhang, Q.; Pan, L.; Wang, P.; Liu, Z.B.; Jiang, W.S.; Chen, Y.; Tian, J.G. Ultrasensitive Flow Sensing of a Single Cell Using Graphene-Based Optical Sensors. Nano Lett. 2014, 14, 3563–3569. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meng, B.; Li, X.; Liang, G.; Hu, X.; Wang, Q.J. Broadband high photoresponse from pure monolayer graphene photodetector. Nat. Commun. 2013, 4, 1811. [Google Scholar] [CrossRef]
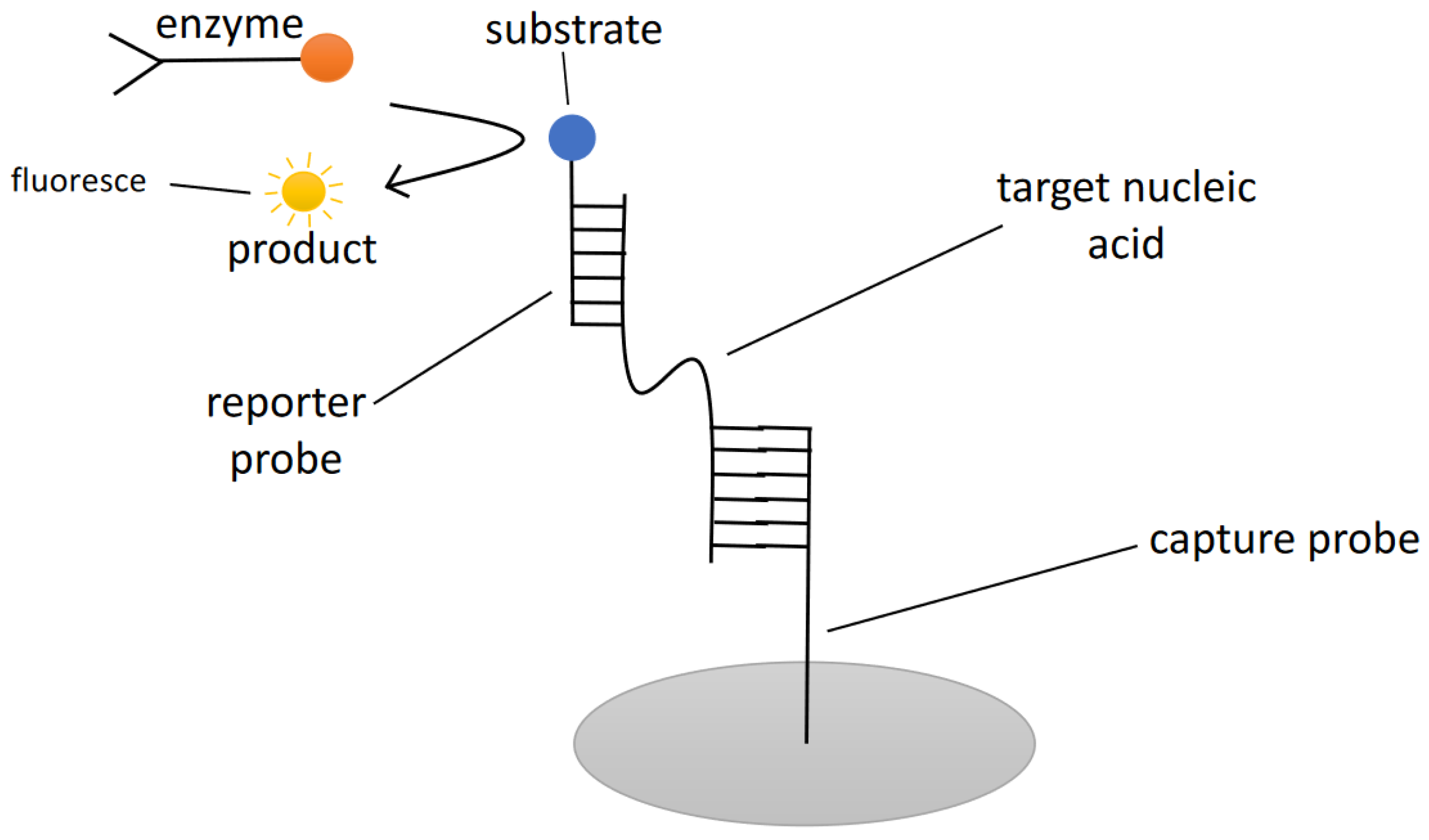

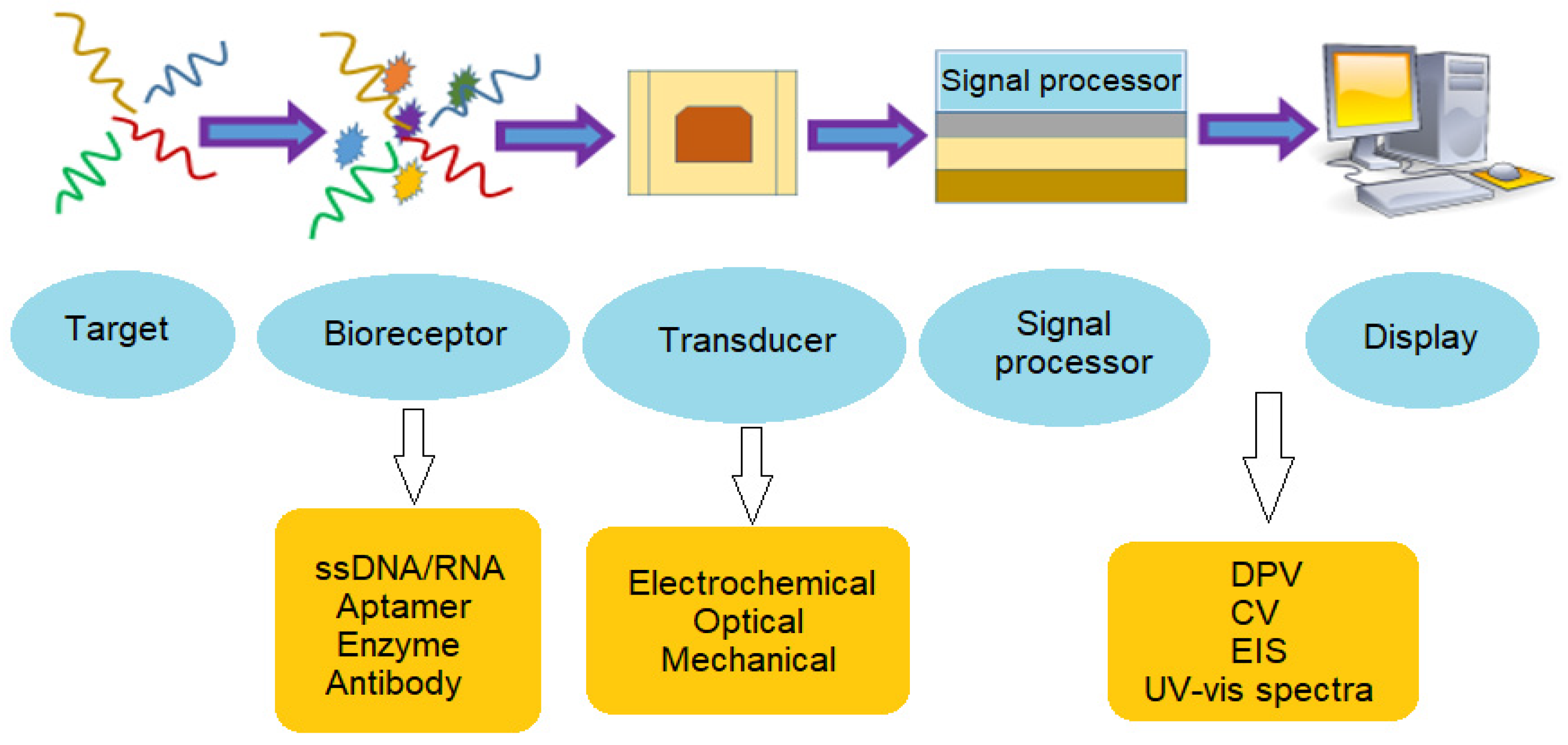

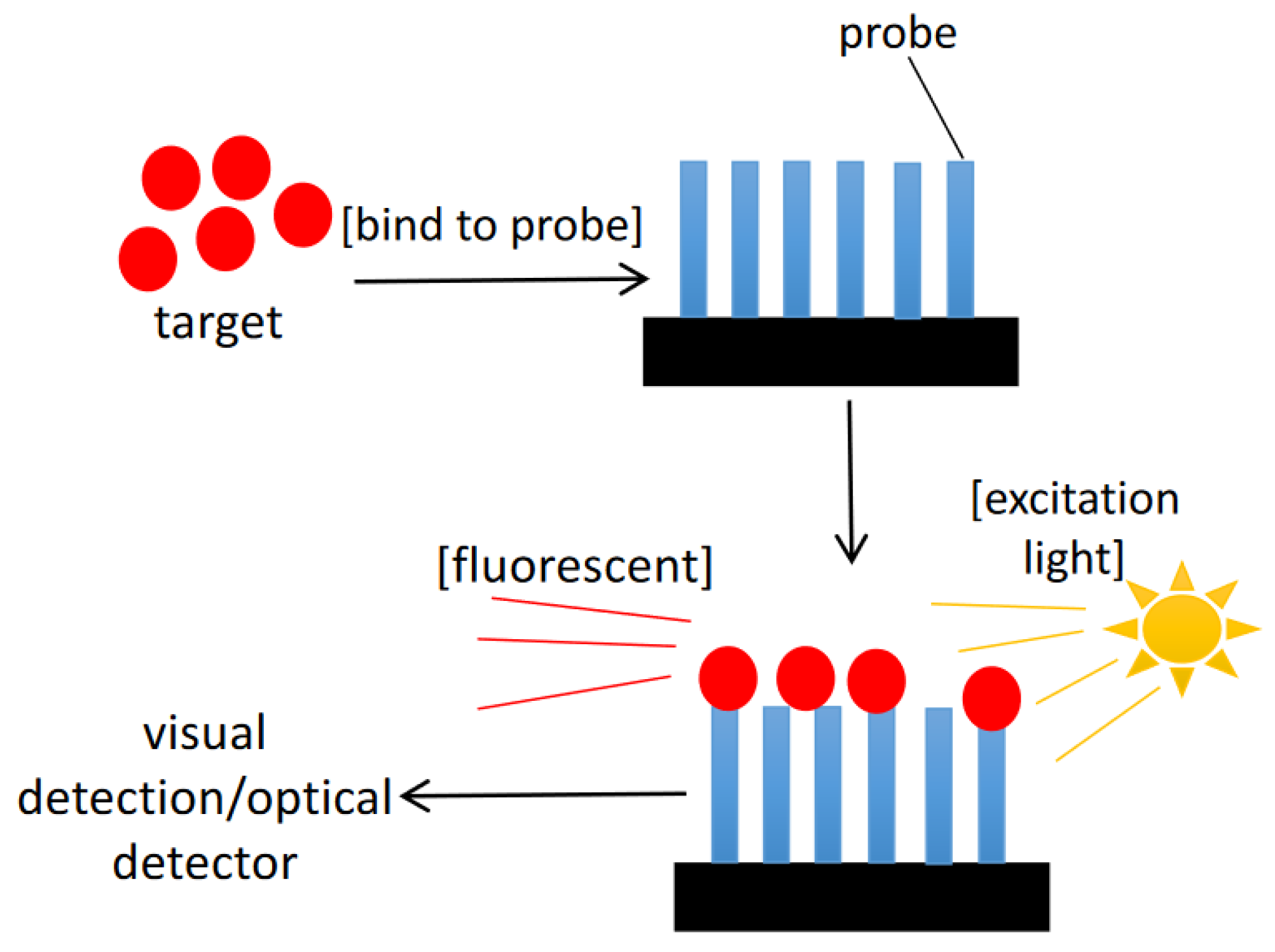
| Reference | Target | Instruments/Methods | Response Time | Detection Limit | Advantages | Drawbacks |
|---|---|---|---|---|---|---|
| [11,36] | Autotrophs | Light Microscopy | 2 h–4 days | - | Useful for the studies of taxanomy and morphology of whole HAB cells. | Incapable of distinguishing cells below 5 μm in size. Possible human errors in counting cells. |
| [36] | Autotrophs | TEM and SEM | 1–10 days | - | Allows identification of pico- and nano-sized organisms. | Time-consuming, expensive. |
| [44] | Prorocentrum donghaiense | FISH | 1 h | - | An accurate detection method. Usage of PNA probe displayed more intensive green fluorescence than DNA probe. | The quantification requires LM and is thereby slow to produce results and possibly comes with human error in counting. |
| [43] | Alexandrium minutum | FISH | 45 min | - | A rapid detection tool for A. minutum | Quantification requires LM. Auto-fluorescence leads to false positive results. Challenge in fixing microalgae cells. |
| [45] | Karlodinium veneficum (Kv), Chattonella marina (Cm), Skeletonema spp., Scrippsiella trochoidea (St), Karenia mikimotoi (Km), and Prorocentrum donghaiense (Pd) | Multiplex PCR (mPCR) | - | 6 cells per reaction (Skeletonema spp., Cm, and St) 60 cells per reaction (Km, Pd, and Kv) | Allows simultaneous multiple targets detection. Strong specificity and stability. | Numerous optimization steps are required to have a good mPCR performance. Lack of accuracy in determining cell density. |
| [49] | Alexandrium spp. | qPCR | - | <5 × 103 cells L−1 | High sensitivity of detection. | Overestimation of abundance. |
| [23] | A. minutum | ELISA | <3 h | 1 × 104 cells sample−1 | Capable of detecting A. minutum cells at different cell counts in the presence of a complex background. | Require at least 10,000 cells for measurable RNA concentration, based on the extraction kit used in this experiment. Enumeration of cell count is based on estimation only. |
| [50] | A. minutum species | Whole-cell ELISA | - | 1 × 105 cells L−1 | Good sensitivity and specificity on natural seawater samples. | Generation of monoclonal antibodies via rats. Tends to overestimate the number of cells by a rough factor of 10. |
| [51] | A. fundyense, Pseudo-nitzschia australis, and A. Ostenfeldii | Fiber optic microarrays | - | 5–10 cells sample−1 (A. fundyense and Pseudo-nitzschia Australis) 50 cells sample−1 (A. Ostenfeldii) | Simultaneous detection of all three species. Simple and reusable sensor with no loss of sensitivity. | Complex instrumentations with microscopic epifluorescence and image analysis. High experimental and set-up costs. |
| [52] | A. fundyense, Pseudo-nitzschia australis, and A. Ostenfeldii | Fiber optic microarrays | 45 min | 5 cells sample−1 | Simultaneous detection with no cross-reactivity. | High experimental and set-up cost. |
| Reference | Target | Instruments/Methods | Response Time (h) | Detection Limit (Cells L−1) | Advantages | Drawbacks |
|---|---|---|---|---|---|---|
| [62] | DNA/RNA of microbial pathogens | Rapid PCR-Detect and Hybrid PCR-Detect. | 4–6 | - | Sensitive detection of sample DNA/RNA. | Only capable of detecting single-base mutations from pure culture isolate. |
| [64] | rRNA of toxic algae (toxic dinoflagellate A. Ostenfeldii) | Molecular DNA probes | 7–10 | 5 × 109 | Simplified detection methods | Manual RNA isolation and manipulation of the hybridization steps are required at high temperature system. |
| [63] | Microbial pathogens and Karenia brevis | 8-plex assay of microbes | 3–5 | 1000 | Able to multi-target electrochemical detection of microbial pathogens. | Complex steps in DNA extraction. |
| [23] | rRNA of harmful algae species | Multi-probe chip and a semi-automated rRNA biosensor | ~2 | - | Allows in situ detection and monitoring of toxic algae. | Manual rRNA isolation. |
| [35] | Alexandrium minutum | Multi-probe biosensor (ALGADEC) | ~2 | 25,000 | Almost fully automated device for in situ analysis. | Poor limit detection. |
| [65] | Prymnesium parvum, Gymnodinium catenatum, and Pseudo-nitzschia australis | DIG-enzymatic label assay | 1–2 | - | Simple and easy handling amperometric techniques. | Very poor response for cyclic voltammetry. |
| [66] | Alexandrium species | Surface Plasmon Resonance (SPR) biosensing instrument and peptide nucleic acid probes | >3.5 | - | Cost effective and yield quick result. | Require tubing flushing maintenance. |
| Reference | Detection Method | Detection Time (min) | Linear Range (Cells L−1) | Detection Limit (Cells L−1) |
|---|---|---|---|---|
| [72] | Electrochemical nanobody immunosensor | ≤75 | 5.00 × 103–1.00 × 109 | 3.1 × 103 |
| [86] | AuNPs-based Immunosensor | ≤30 | - | 1 × 103 |
| [90] | Loop-mediated isothermal amplification assay | ≤120 | ≈1.00 × 104–1.00 × 108 | ≈1.7 × 104 |
| [91] | Quartz crystal microbalance | ≤80 | 1.50 × 109–5.50 × 109 | 1 × 109 |
| [92] | Super-paramagnetic immunochromatographic strip test | ≤30 | ≈2.00 × 105–2.45 × 108 | 5 × 104 |
| Methods | Advantages | Drawbacks | * Detection Time | Portablity | Ease of Operation |
|---|---|---|---|---|---|
| Microscopic assay | Useful for taxonomy and morphology study | Time consuming and expensive | Very slow | Lab based | Fairly easy |
| FISH | Accurate detection method | Quantification requires a microscopy approach | Moderate | Lab based | Fairly easy |
| PCR | High specificity of detection | Numerous optimization steps are required for good test results | Slow/Moderate | Lab based | Complicated |
| ELISA | Good specificity even in the presence of complex background | Expensive | Slow | Lab based | Complicated |
| Microarray | Allows numerous simultaneous detections, good sensitivity | Expensive | Moderate | Lab based | Complicated |
| Electrochemical Biosensor | Small, simple, robust devices, and good detection limits | Device less reproducible | Fast | On-site | Easy |
| Optical Biosensor | High specificity and cost-effective | Device less reproducible | Fast | On-site | Easy |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chin Chwan Chuong, J.J.; Rahman, M.; Ibrahim, N.; Heng, L.Y.; Tan, L.L.; Ahmad, A. Harmful Microalgae Detection: Biosensors versus Some Conventional Methods. Sensors 2022, 22, 3144. https://doi.org/10.3390/s22093144
Chin Chwan Chuong JJ, Rahman M, Ibrahim N, Heng LY, Tan LL, Ahmad A. Harmful Microalgae Detection: Biosensors versus Some Conventional Methods. Sensors. 2022; 22(9):3144. https://doi.org/10.3390/s22093144
Chicago/Turabian StyleChin Chwan Chuong, Jeremy Jason, Mahbubur Rahman, Nadiah Ibrahim, Lee Yook Heng, Ling Ling Tan, and Asmat Ahmad. 2022. "Harmful Microalgae Detection: Biosensors versus Some Conventional Methods" Sensors 22, no. 9: 3144. https://doi.org/10.3390/s22093144
APA StyleChin Chwan Chuong, J. J., Rahman, M., Ibrahim, N., Heng, L. Y., Tan, L. L., & Ahmad, A. (2022). Harmful Microalgae Detection: Biosensors versus Some Conventional Methods. Sensors, 22(9), 3144. https://doi.org/10.3390/s22093144







