Fluorescence DNA Switch for Highly Sensitive Detection of miRNA Amplified by Duplex-Specific Nuclease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Preparation of DNA Modified AuNPs
2.3. Preparation of Gold Electrode
2.4. DSN-Mediated Amplification and Dumbbell DNA Switch
2.5. Electrochemical Measurements
2.6. Fluorescent Measurements
2.7. UV-vis Measurements and Polyacrylamide Gel Electrophoresis (PAGE)
3. Results and Discussion
3.1. Sensing Principle
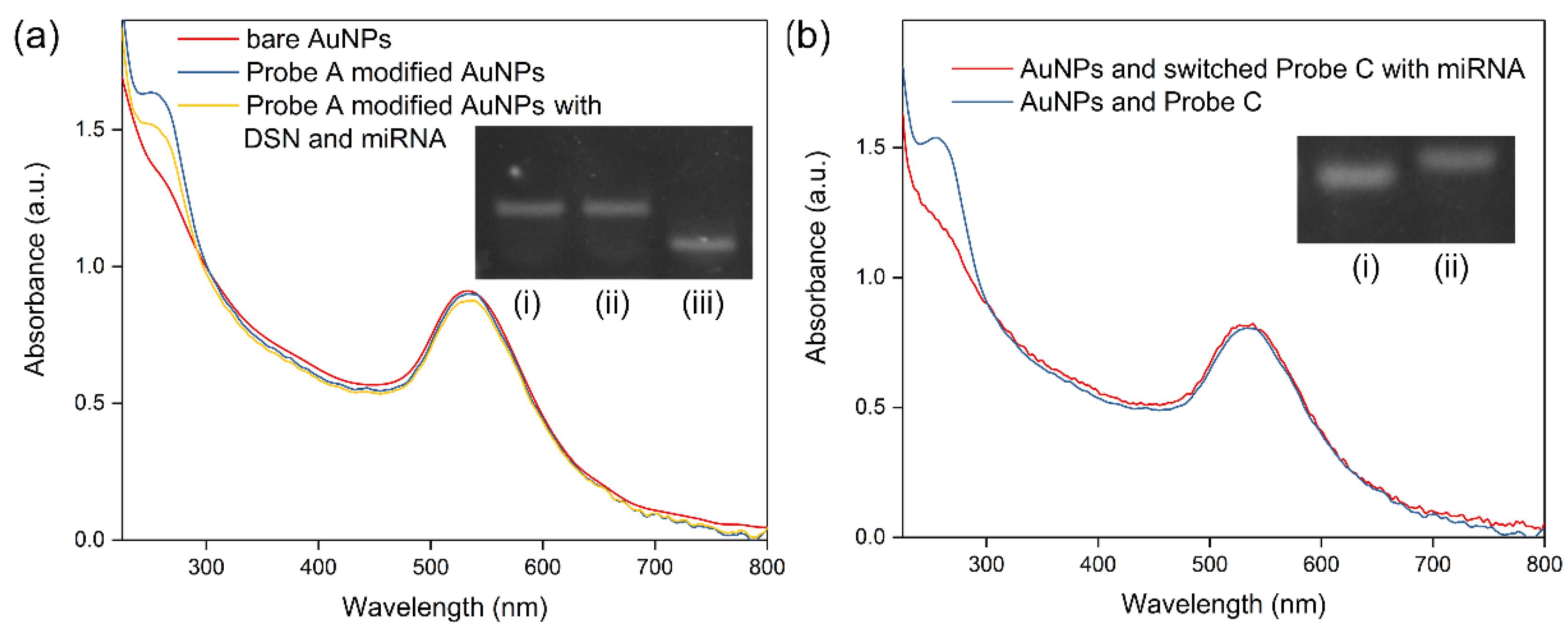
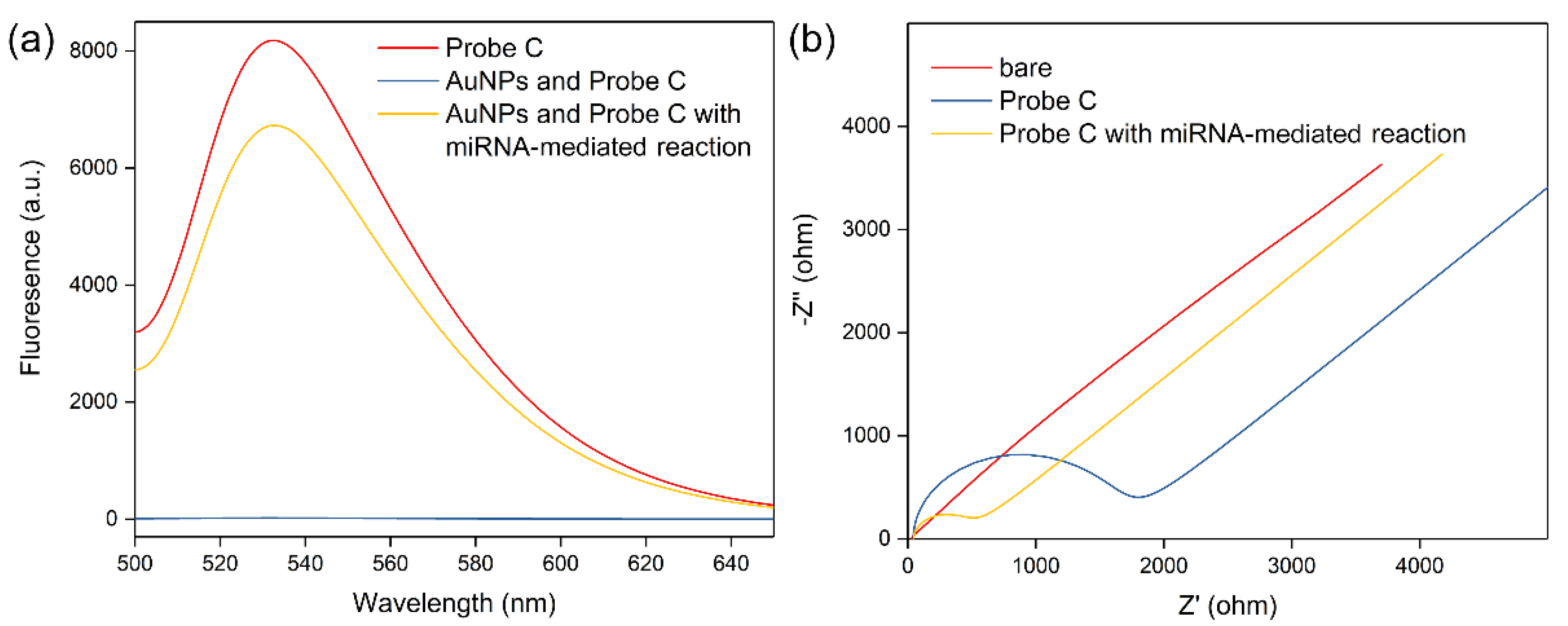
3.2. Feasibility of the Working Mechanism
3.3. Quantification of miRNA
3.4. Investigations of Selectivity and Practical Utility
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garcia-Martin, R.; Wang, G.X.; Brandão, B.B.; Zanotto, T.M.; Shah, S.; Patel, S.K.; Schilling, B.; Kahn, C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2022, 601, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Sun, X.D.; Hu, S.; Wang, M.Q.; Ma, N.; Chen, J.H.; Duan, F. Long noncoding RNA SNHG1 silencing accelerates hepatocyte-like cell differentiation of bone marrow-derived mesenchymal stem cells to alleviate cirrhosis via the microRNA-15a/SMURF1/UVRAG axis. Cell Death Discov. 2022, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.L.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, J.; Li, M.Q.; Xu, J.; Zhang, J.P.; Jin, L.P. MicroRNA-184 promotes apoptosis of trophoblast cells via targeting WIG1 and induces early spontaneous abortion. Cell Death Dis. 2019, 10, 223. [Google Scholar] [CrossRef]
- Lempriere, S. Exosomal microRNA is promising biomarker in PD. Nat. Rev. Neurol. 2022, 18, 65. [Google Scholar] [CrossRef]
- Raut, J.R.; Schöttker, B.; Holleczek, B.; Guo, F.; Bhardwaj, M.; Miah, K.; Schrotz-King, P.; Brenner, H. A microRNA panel compared to environmental and polygenic scores for colorectal cancer risk prediction. Nat. Commun. 2021, 12, 4811. [Google Scholar] [CrossRef]
- Bockhorn, J.; Dalton, R.; Nwachukwu, C.; Huang, S.; Prat, A.; Yee, K.; Chang, Y.F.; Huo, D.Z.; Wen, Y.J.; Swanson, K.E.; et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat. Commun. 2013, 4, 1393. [Google Scholar] [CrossRef] [Green Version]
- Bauer, K.M.; Hummon, A.B. Effects of the miR-143/-145 microRNA cluster on the colon cancer proteome and transcriptome. J. Proteome Res. 2012, 11, 4744–4754. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.Y.; Jolly, M.K.; Gomoto, R.; Huang, B.; Onuchic, J.; Ben-Jacob, E. Tristability in cancer-associated microRNA-TF chimera toggle switch. J. Phys. Chem. B 2013, 117, 13164–13174. [Google Scholar] [CrossRef]
- Thomson, J.M.; Parker, J.; Perou, C.M.; Hammond, S.M. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods 2004, 1, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Park, J.-E.; Nam, J.-M. Bio-barcode gel assay for microRNA. Nat. Commun. 2014, 5, 3367. [Google Scholar] [CrossRef] [PubMed]
- Haenisch, S.; von Rüden, E.-L.; Wahmkow, H.; Rettenbeck, M.L.; Michler, C.; Russmann, V.; Bruckmueller, H.; Waetzig, V.; Cascorbi, I.; Potschka, H. miRNA-187-3p-mediated regulation of the KCNK10/TREK-2 potassium channel in a rat epilepsy model. ACS Chem. Neurosci. 2016, 7, 1585–1594. [Google Scholar] [CrossRef] [PubMed]
- Xian, L.M.; Xu, F.; Liu, J.Z.; Xu, N.; Li, H.D.; Ge, H.Y.; Shao, K.; Fan, J.L.; Xiao, G.S.; Peng, X.J. MicroRNA detection with turnover amplification via hybridization-mediated staudinger reduction for pancreatic cancer diagnosis. J. Am. Chem. Soc. 2019, 141, 20490–20497. [Google Scholar] [CrossRef] [PubMed]
- Miao, P.; Tang, Y.G. Dumbbell hybridization chain reaction based electrochemical biosensor for ultrasensitive detection of exosomal miRNA. Anal. Chem. 2020, 92, 12026–12032. [Google Scholar] [CrossRef] [PubMed]
- Song, S.C.; Li, N.; Bai, L.P.; Gai, P.P.; Li, F. Photo-assisted robust anti-interference self-powered biosensing of microRNA based on Pt–S bonds and the inorganic–organic hybridization strategy. Anal. Chem. 2022, 94, 1654–1660. [Google Scholar] [CrossRef]
- Gutiérrez-Gálvez, L.; García-Mendiola, T.; Gutiérrez-Sánchez, C.; Guerrero-Esteban, T.; García-Diego, C.; Buendía, I.; García-Bermejo, M.L.; Pariente, F.; Lorenzo, E. Carbon nanodot–based electrogenerated chemiluminescence biosensor for miRNA-21 detection. Microchim. Acta 2021, 188, 398. [Google Scholar] [CrossRef]
- Miao, P.; Tang, Y.G. Two-dimensional hybridization chain reaction strategy for highly sensitive analysis of intracellular mRNA. Anal. Chem. 2020, 92, 12700–12709. [Google Scholar] [CrossRef]
- Prusty, D.K.; Adam, V.; Zadegan, R.M.; Irsen, S.; Famulok, M. Supramolecular aptamer nano-constructs for receptor-mediated targeting and light-triggered release of chemotherapeutics into cancer cells. Nat. Commun. 2018, 9, 535. [Google Scholar] [CrossRef] [Green Version]
- Aw, S.S.; Tang, M.X.; Teo, Y.N.; Cohen, S.M. A conformation-induced fluorescence method for microRNA detection. Nucleic Acids Res. 2016, 44, e92. [Google Scholar] [CrossRef] [Green Version]
- Tavallaie, R.; McCarroll, J.; Le Grand, M.; Ariotti, N.; Schuhmann, W.; Bakker, E.; Tilley, R.D.; Hibbert, D.B.; Kavallaris, M.; Gooding, J.J. Nucleic acid hybridization on an electrically reconfigurable network of gold-coated magnetic nanoparticles enables microRNA detection in blood. Nat. Nanotechnol. 2018, 13, 1066–1071. [Google Scholar] [CrossRef]
- Miao, P.; Tang, Y.G. DNA walking and rolling nanomachine for electrochemical detection of miRNA. Small 2020, 16, 2004518. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.C.; Liu, Y.Q.; Ye, B.C. One-step, multiplexed fluorescence detection of microRNAs based on duplex-specific nuclease signal amplification. J. Am. Chem. Soc. 2012, 134, 5064–5067. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.S.; Man, T.T.; Zhu, C.F.; Pei, H.; Shi, J.Y.; Li, L.; Qu, X.M.; Shen, X.Z.; Li, J. MoS2 nanoprobe for microRNA quantification based on duplex-specific nuclease signal amplification. ACS Appl. Mater. Interfaces 2018, 10, 7852–7858. [Google Scholar] [CrossRef] [PubMed]
- Dulkeith, E.; Ringler, M.; Klar, T.A.; Feldmann, J.; Javier, A.M.; Parak, W.J. Gold nanoparticles quench fluorescence by phase induced radiative rate suppression. Nano Lett. 2005, 5, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Bo, B.; Zhang, T.; Jiang, Y.T.; Cui, H.Y.; Miao, P. Triple signal amplification strategy for ultrasensitive determination of miRNA based on duplex specific nuclease and bridge DNA–gold nanoparticles. Anal. Chem. 2018, 90, 2395–2400. [Google Scholar] [CrossRef]
- Zhang, X.; Servos, M.R.; Liu, J.W. Instantaneous and Quantitative Functionalization of Gold Nanoparticles with Thiolated DNA Using a pH-Assisted and Surfactant-Free Route. J. Am. Chem. Soc. 2012, 134, 7266–7269. [Google Scholar] [CrossRef] [Green Version]
- Samanta, A.; Zhou, Y.D.; Zou, S.L.; Yan, H.; Liu, Y. Fluorescence quenching of quantum dots by gold nanoparticles: A potential long range spectroscopic ruler. Nano Lett. 2014, 14, 5052–5057. [Google Scholar] [CrossRef]
- Yue, S.Z.; Song, X.Y.; Song, W.L.; Bi, S. An enzyme-free molecular catalytic device: Dynamically self-assembled DNA dendrimers for in situ imaging of microRNAs in live cells. Chem. Sci. 2019, 10, 1651–1658. [Google Scholar] [CrossRef] [Green Version]
- Ying, Z.M.; Wu, Z.; Tu, B.; Tan, W.H.; Jiang, J.H. Genetically encoded fluorescent RNA sensor for ratiometric imaging of microRNA in living tumor cells. J. Am. Chem. Soc. 2017, 139, 9779–9782. [Google Scholar] [CrossRef]
- Si, Y.M.; Xu, L.; Wang, N.N.; Zheng, J.; Yang, R.H.; Li, J.S. Target microRNA-responsive DNA hydrogel-based surface-enhanced raman scattering sensor arrays for microRNA-marked cancer screening. Anal. Chem. 2020, 92, 2649–2655. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, T.; Li, J.; Gong, H.; Chen, C.Y.; Chen, X.M.; Cai, C.Q. Ratiometric fluorescence sensor for the microRNA determination by catalyzed hairpin assembly. ACS Sens. 2017, 2, 1430–1434. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.X.; Wu, R.; Fan, G.C.; Deng, A.P.; Zhu, J.J. Enzyme-free photoelectrochemical biosensor based on the co-sensitization effect coupled with dual cascade toehold-mediated strand displacement amplification for the sensitive detection of microRNA-21. ACS Sustain. Chem. Eng. 2018, 6, 11633–11641. [Google Scholar] [CrossRef]
- Qian, S.Y.; Lin, M.; Ji, W.; Yuan, H.Z.; Zhang, Y.; Jing, Z.G.; Zhao, J.Z.; Masson, J.F.; Peng, W. Boronic acid functionalized Au nanoparticles for selective microRNA signal amplification in fiber-optic surface plasmon resonance sensing system. ACS Sens. 2018, 3, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Deng, W.F.; Tan, Y.M.; Xie, Q.J. A glucose/O2 biofuel cell integrated with an exonuclease-powered DNA walker for self-powered sensing of microRNA. Chem. Commun. 2022, 58, 2922–2925. [Google Scholar] [CrossRef]
- Miao, P.; Tang, Y.G. Cascade toehold-mediated strand displacement reaction for ultrasensitive detection of exosomal microRNA. CCS Chem. 2020, 2, 2331–2339. [Google Scholar] [CrossRef]




| Name | Sequence (from 5′ to 3′) |
|---|---|
| Probe A | GTTGGAGCTAGTGGCGTAGAACTATACAACCTACTACCTCA-(CH2)6-SH |
| Probe B | GTTGGAGCTAGTGGCGTAG |
| Probe C | SH-(CH2)3-TCCAACGGCGTAGTCGAAATAGGCTACGCCACTAGCTCCAACTCGAAATAGGGTTGGATTT-Alexa Fluor 488 |
| hsa-let-7a | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7b | UGAGGUAGUAGGUUGUGUGGU |
| hsa-let-7c | UGAGGUAGUAGGUUGUAUGGUU |
| hsa-let-7d | AGAGGUAGUAGGUUGCAUAGUU |
| hsa-let-7e | UGAGGUAGGAGGUUGUAUAGUU |
| hsa-let-7f | UGAGGUAGUAGAUUGUAUAGUU |
| hsa-let-7g | UGAGGUAGUAGUUUGUACAGUU |
| Technique | Strategy | LOD (M) | Ref |
|---|---|---|---|
| fluorescence | self-assembled DNA dendrimers | 2.5 × 10−9 | [28] |
| fluorescence | light-up RNA aptamers | 3 × 10−10 | [29] |
| surface-enhanced Raman scattering | target responsive DNA hydrogel | 1.1 × 10−10 | [30] |
| fluorescence | catalyzed hairpin assembly based ratiometric assay | 7.2 × 10−11 | [31] |
| photoelectrochemistry | dual cascade toehold-mediated strand displacement amplification | 3.1 × 10−13 | [32] |
| surface plasmon resonance | boronic acid functionalized AuNPs | 2.7 × 10−13 | [33] |
| biofuel cells | exonuclease-powered DNA walker | 4.5 × 10−17 | [34] |
| square wave voltammetry | cascade toehold-mediated strand displacement reaction | 4 × 10−17 | [35] |
| fluorescence | dumbbell DNA switch and DSN-mediated amplification | 10−16 | this |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Guo, Z.; Luo, G.; Miao, P. Fluorescence DNA Switch for Highly Sensitive Detection of miRNA Amplified by Duplex-Specific Nuclease. Sensors 2022, 22, 3252. https://doi.org/10.3390/s22093252
Li X, Guo Z, Luo G, Miao P. Fluorescence DNA Switch for Highly Sensitive Detection of miRNA Amplified by Duplex-Specific Nuclease. Sensors. 2022; 22(9):3252. https://doi.org/10.3390/s22093252
Chicago/Turabian StyleLi, Xiaoqiang, Zhenzhen Guo, Gangyin Luo, and Peng Miao. 2022. "Fluorescence DNA Switch for Highly Sensitive Detection of miRNA Amplified by Duplex-Specific Nuclease" Sensors 22, no. 9: 3252. https://doi.org/10.3390/s22093252
APA StyleLi, X., Guo, Z., Luo, G., & Miao, P. (2022). Fluorescence DNA Switch for Highly Sensitive Detection of miRNA Amplified by Duplex-Specific Nuclease. Sensors, 22(9), 3252. https://doi.org/10.3390/s22093252






