Plant Root Phenotyping Using Deep Conditional GANs and Binary Semantic Segmentation
Abstract
1. Introduction
2. Background
3. Methodology
3.1. Dataset Acquisition
3.2. Semantic Map Creation
3.3. cGAN Model Selection and Training
3.4. Semantic Segmentation Model Selection and Training
3.5. Segmentation Postprocessing
3.6. Evaluation Metrics
3.7. Model Comparison
3.8. Summary of Methodology
4. Experimental Results
4.1. Pix2PixHD Results
4.2. SegNet Results
4.3. Postprocessing Results
4.4. Model Comparison Results
5. Future Work
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GAN | Generative Adversarial Network |
| cGAN | Conditional Generative Adversarial Network |
| UGV | Unmanned Ground Vehicle |
| MRI | Magnetic Resonance Imaging |
| CNN | Convolutional Neural Networks |
| RSA | Root System Architecture |
| PIL | Python Image Library |
| IOU | Intersection Over Union |
| TP | True Positive |
| FP | False Positive |
| TN | True Negative |
| FN | False Negative |
| ELU | Exponential Linear Unit |
| ResUNet | Residual UNet |
| DSResUNet | Deeply Supervised Residual UNet |
References
- Gong, L.; Du, X.; Zhu, K.; Lin, C.; Lin, K.; Wang, T.; Lou, Q.; Yuan, Z.; Huang, G.; Liu, C. Pixel level segmentation of early-stage in-bag rice root for its architecture analysis. Comput. Electron. Agric. 2021, 186, 106197. [Google Scholar] [CrossRef]
- Wang, T.; Rostamza, M.; Song, Z.; Wang, L.; McNickle, G.; Iyer-Pascuzzi, A.S.; Qiu, Z.; Jin, J. SegRoot: A high throughput segmentation method for root image analysis. Comput. Electron. Agric. 2019, 162, 845–854. [Google Scholar] [CrossRef]
- Gaggion, N.; Ariel, F.; Daric, V.; Lambert, É.; Legendre, S.; Roulé, T.; Camoirano, A.; Milone, D.; Crespi, M.; Blein, T.; et al. ChronoRoot: High-throughput phenotyping by deep segmentation networks reveals novel temporal parameters of plant root system architecture. GigaScience 2021, 10, giab052. [Google Scholar] [PubMed]
- Bucksch, A.; Burridge, J.; York, L.M.; Das, A.; Nord, E.; Weitz, J.S.; Lynch, J.P. Image-based high-throughput field phenotyping of crop roots. Plant Physiol. 2014, 166, 470–486. [Google Scholar] [CrossRef]
- Smith, A.G.; Petersen, J.; Selvan, R.; Rasmussen, C.R. Segmentation of roots in soil with U-Net. Plant Methods 2020, 16, 1–15. [Google Scholar] [CrossRef]
- Jez, J.M.; Topp, C.N.; Buckner, E.; Tong, H.; Ottley, C.; Williams, C. High-throughput image segmentation and machine learning approaches in the plant sciences across multiple scales. Emerg. Top. Life Sci. 2021, 5, 239–248. [Google Scholar] [CrossRef]
- Shen, C.; Liu, L.; Zhu, L.; Kang, J.; Wang, N.; Shao, L. High-throughput in situ root image segmentation based on the improved DeepLabv3+ method. Front. Plant Sci. 2020, 11, 576791. [Google Scholar] [CrossRef]
- Mairhofer, S.; Zappala, S.; Tracy, S.; Sturrock, C.; Bennett, M.J.; Mooney, S.J.; Pridmore, T.P. Recovering complete plant root system architectures from soil via X-ray μ-computed tomography. Plant Methods 2013, 9, 1–7. [Google Scholar] [CrossRef]
- Wang, T.C.; Liu, M.Y.; Zhu, J.Y.; Tao, A.; Kautz, J.; Catanzaro, B. High-Resolution Image Synthesis and Semantic Manipulation with Conditional GANs. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Yasrab, R.; Zhang, J.; Smyth, P.; Pound, M.P. Predicting plant growth from time-series data using deep learning. Remote Sens. 2021, 13, 331. [Google Scholar] [CrossRef]
- Karras, T.; Aila, T.; Laine, S.; Lehtinen, J. Progressive growing of gans for improved quality, stability, and variation. arXiv 2017, arXiv:1710.10196. [Google Scholar]
- Möller, B.; Schreck, B.; Posch, S. Analysis of Arabidopsis Root Images–Studies on CNNs and Skeleton-Based Root Topology. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 1294–1302. [Google Scholar]
- Pattanayak, D.; Patel, K. Generative Adversarial Networks: Solution for Handling Imbalanced Datasets in Computer Vision. In Proceedings of the IEEE 2022 International Conference for Advancement in Technology (ICONAT), Goa, India, 21–22 January 2022. [Google Scholar]
- Jiang, Y.; Li, C. Convolutional neural networks for image-based high-throughput plant phenotyping: A review. Plant Phenom. 2020, 2020, 4152816. [Google Scholar] [CrossRef] [PubMed]
- Sampath, V.; Maurtua, I.; Aguilar Martín, J.J.; Gutierrez, A. A survey on generative adversarial networks for imbalance problems in computer vision tasks. J. Big Data 2021, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Atanbori, J.; Montoya-P, M.E.; Selvaraj, M.G.; French, A.P.; Pridmore, T.P. Convolutional neural net-based cassava storage root counting using real and synthetic images. Front. Plant Sci. 2019, 10, 1516. [Google Scholar] [CrossRef] [PubMed]
- Mi, J.; Gao, W.; Yang, S.; Hao, X.; Li, M.; Wang, M.; Zheng, L. A method of plant root image restoration based on GAN. IFAC-PapersOnLine 2019, 52, 219–224. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Pang, Y.; Lin, J.; Qin, T.; Chen, Z. Image-to-image translation: Methods and applications. IEEE Trans. Multimed. 2021. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Cody Hazlett, H.; Gimpel Smith, R.; Ho, S.; Gee, J.C.; Gerig, G. User-Guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef]
- Dash, A.; Ye, J.; Wang, G.; Jin, H. High resolution solar image generation using generative adversarial networks. Ann. Data Sci. 2022. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Yeung, M.; Sala, E.; Schönlieb, C.B.; Rundo, L. Unified focal loss: Generalising dice and cross entropy-based losses to handle class imbalanced medical image segmentation. Comput. Med. Imaging Graph. 2022, 95, 102026. [Google Scholar] [CrossRef] [PubMed]
- Buzzy, M.; Thesma, V.; Davoodi, M.; Mohammadpour Velni, J. Real-time plant leaf counting using deep object detection networks. Sensors 2020, 20, 6896. [Google Scholar] [CrossRef] [PubMed]
- Bertels, J.; Eelbode, T.; Berman, M.; Vandermeulen, D.; Maes, F.; Bisschops, R.; Blaschko, M.B. Optimizing the dice score and jaccard index for medical image segmentation: Theory and practice. In Medical Image Computing and Computer-Assisted Intervention. MICCAI 2019; Springer: Cham, Switzerland, 2019; pp. 92–100. [Google Scholar]
- Wasaya, A.; Zhang, X.; Fang, Q.; Yan, Z. Root phenotyping for drought tolerance: A review. Agronomy 2018, 8, 241. [Google Scholar] [CrossRef]

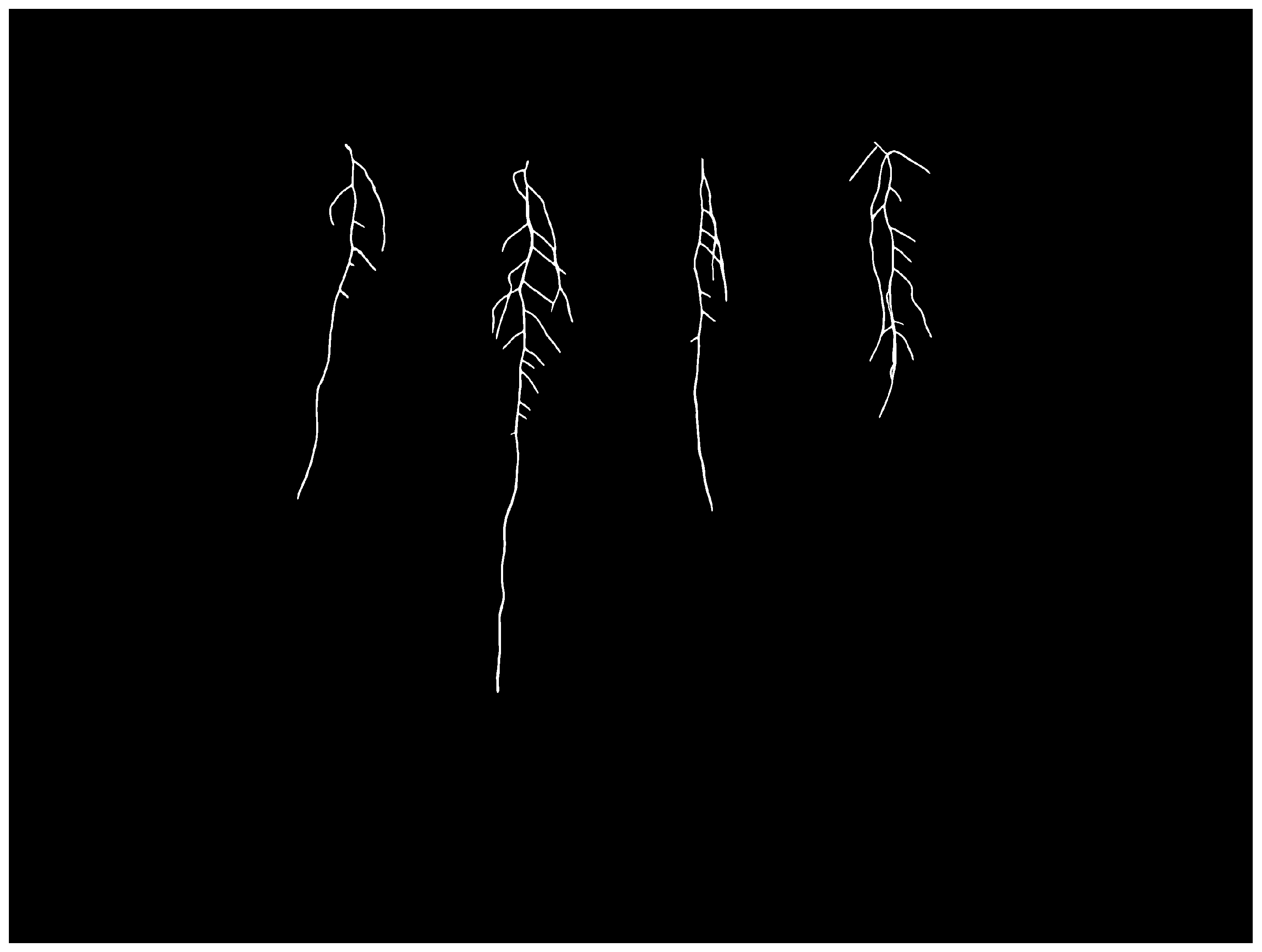










| Metric | SegNet Performance |
|---|---|
| Cross Entropy Loss | 0.020 |
| Accuracy | 0.991 |
| Mean IOU | 65.87% |
| Dice Score | 0.7942 |
| Inference Time | 0.3002 s |
| Metric | SegNet (Ours) | DSResUNet | UNet |
|---|---|---|---|
| Accuracy | 0.991 | 0.994 | 0.995 |
| Mean IOU | 65.87% | 42.78% | 54.34% |
| Dice Score | 0.7942 | 0.5582 | 0.6839 |
| Inference Time | 0.3002 s | 0.2881 s | 0.1728 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thesma, V.; Mohammadpour Velni, J. Plant Root Phenotyping Using Deep Conditional GANs and Binary Semantic Segmentation. Sensors 2023, 23, 309. https://doi.org/10.3390/s23010309
Thesma V, Mohammadpour Velni J. Plant Root Phenotyping Using Deep Conditional GANs and Binary Semantic Segmentation. Sensors. 2023; 23(1):309. https://doi.org/10.3390/s23010309
Chicago/Turabian StyleThesma, Vaishnavi, and Javad Mohammadpour Velni. 2023. "Plant Root Phenotyping Using Deep Conditional GANs and Binary Semantic Segmentation" Sensors 23, no. 1: 309. https://doi.org/10.3390/s23010309
APA StyleThesma, V., & Mohammadpour Velni, J. (2023). Plant Root Phenotyping Using Deep Conditional GANs and Binary Semantic Segmentation. Sensors, 23(1), 309. https://doi.org/10.3390/s23010309





