Prediction of Honeydew Contaminations on Cotton Samples by In-Line UV Hyperspectral Imaging
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Preparation of Solutions
2.2. Sample Set and Sample Preparation
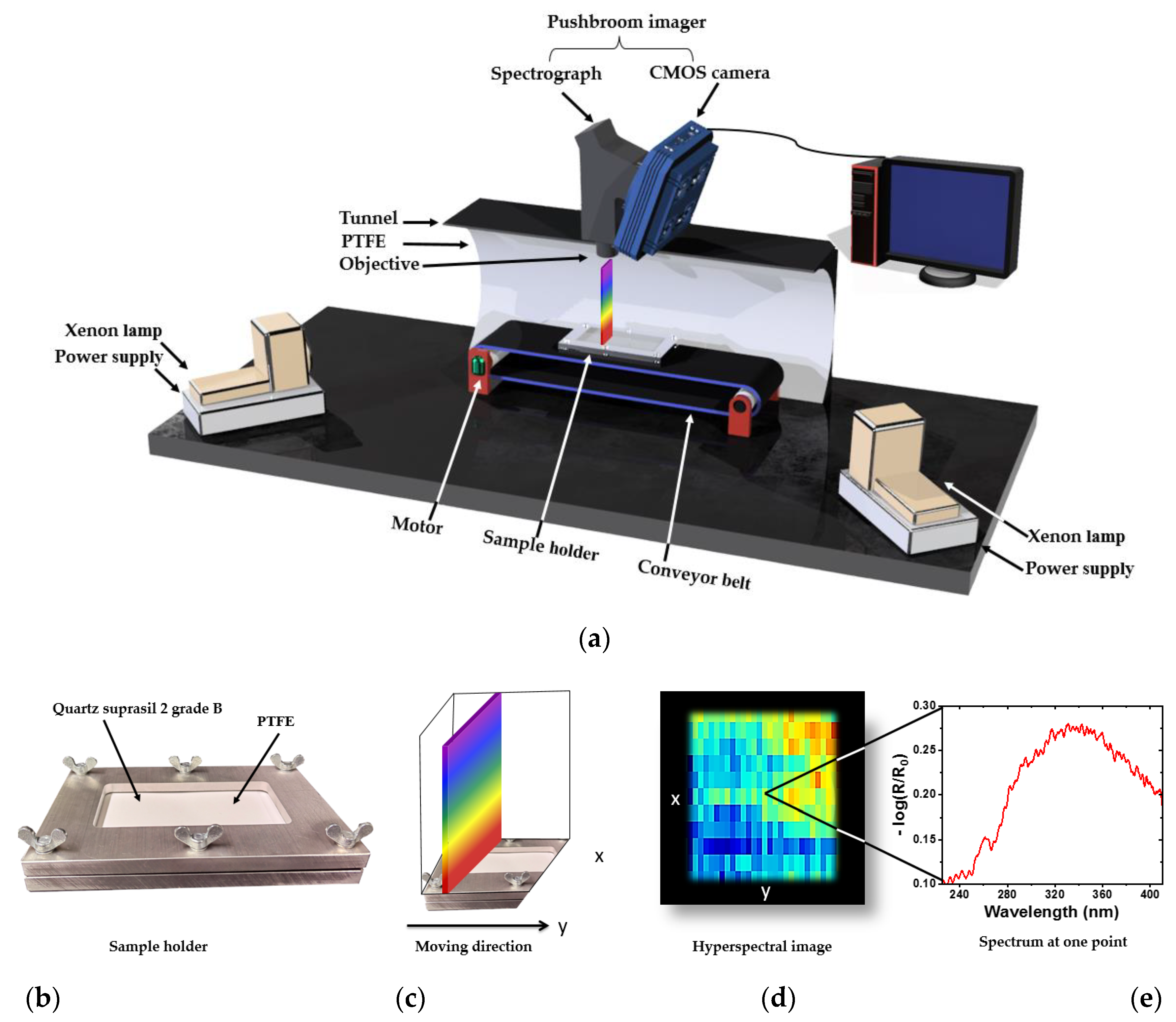
2.3. UV Hyperspectral Imaging Setup
2.4. Data Collection and Preprocessing
2.5. Multivariate Data Analysis and Model Building
3. Results and Discussion
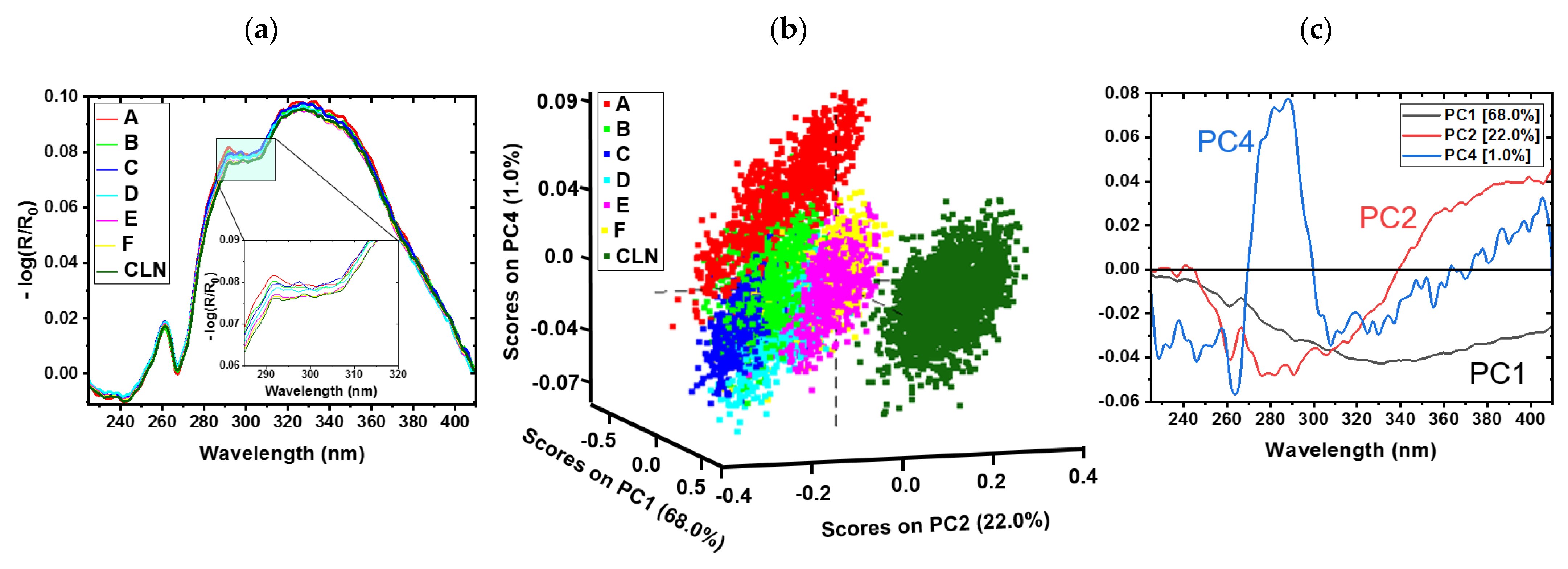
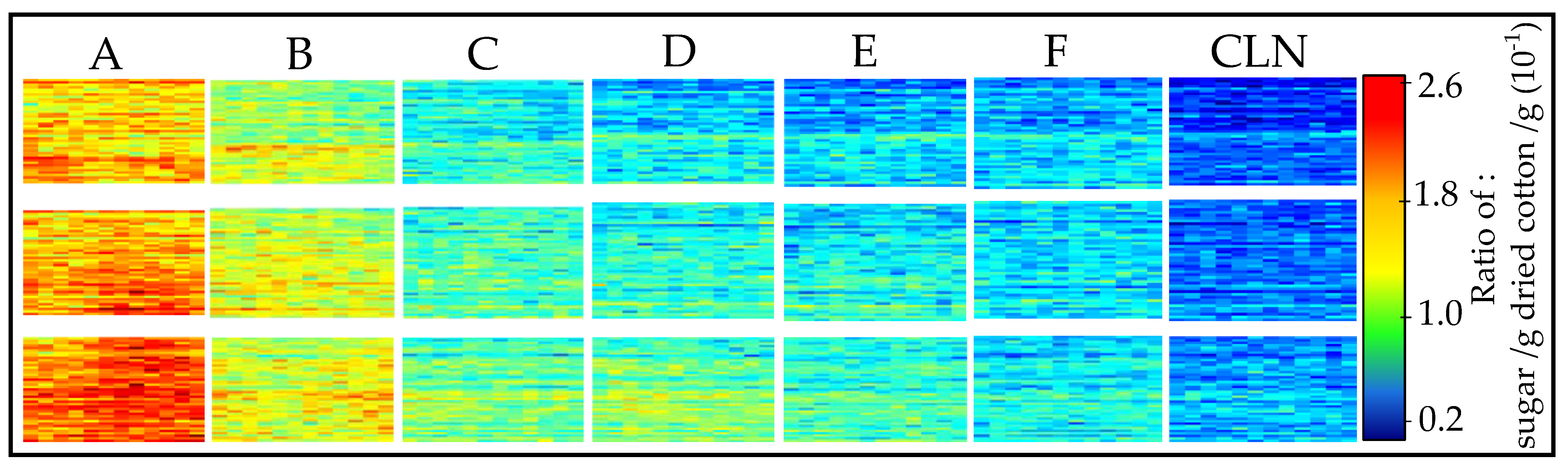
3.1. Cotton Samples Impregnated with Sugar
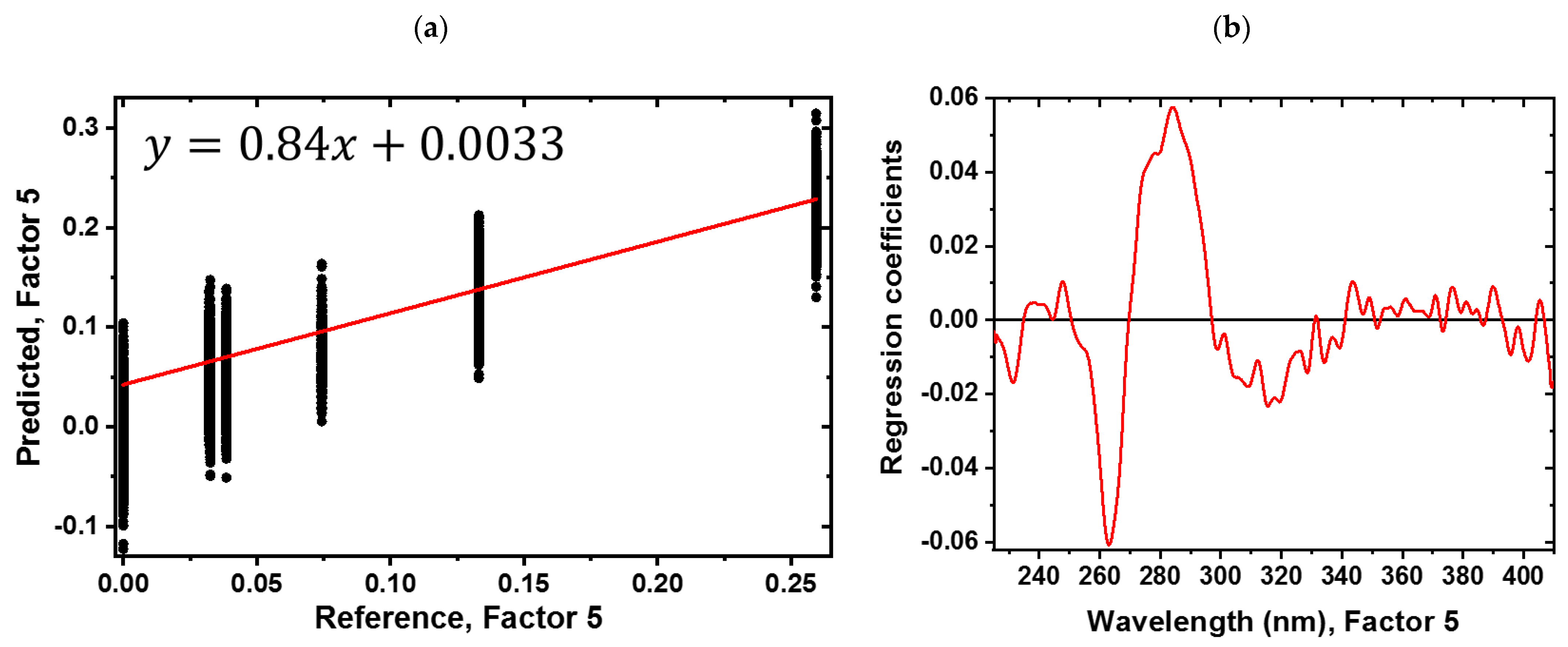
3.2. Predicting the Amount of Sugar and Honeydew Based on the Sugar PLS-R Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Femenias, A.; Marín, S. Hyperspectral Imaging; Vicente, M., Gómez-López, R.B., Eds.; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2021; pp. 363–390. [Google Scholar]
- Al Ktash, M.; Hauler, O.; Ostertag, E.; Brecht, M. Ultraviolet-visible/near infrared spectroscopy and hyperspectral imaging to study the different types of raw cotton. J. Spectr. Imaging 2020, 9, a18. [Google Scholar] [CrossRef]
- Park, B.; Lu, R. Hyperspectral Imaging Technology in Food and Agriculture; Barbosa-Ca’novas, G.V., Ed.; Springer: Athens, GA, USA, 2015. [Google Scholar]
- Gowen, A.A.; O’Donnell, C.P.; Cullen, P.J.; Downey, G.; Frias, J.M. Hyperspectral imaging–an emerging process analytical tool for food quality and safety control. Trends Food Sci. Technol. 2007, 18, 590–598. [Google Scholar] [CrossRef]
- Al Ktash, M.; Stefanakis, M.; Boldrini, B.; Ostertag, E.; Brecht, M. Characterization of pharmaceutical tablets using UV hyperspectral imaging as a rapid in-line analysis tool. Sensors 2021, 21, 4436. [Google Scholar] [CrossRef]
- Al Ktash, M.; Stefanakis, M.; Englert, T.; Drechsel, M.S.; Stiedl, J.; Green, S.; Jacob, T.; Boldrini, B.; Ostertag, E.; Rebner, K. UV hyperspectral imaging as process analytical tool for the characterization of oxide layers and copper states on direct bonded copper. Sensors 2021, 21, 7332. [Google Scholar] [CrossRef]
- Chen, S.-Y.; Chang, C.-Y.; Ou, C.-S.; Lien, C.-T. Detection of insect damage in green coffee beans using VIS-NIR hyperspectral imaging. Remote Sens. 2020, 12, 2348. [Google Scholar] [CrossRef]
- Devassy, B.M.; George, S. Estimation of strawberry firmness using hyperspectral imaging: A comparison of regression models. J. Spectr. Imaging 2021, 10, a3. [Google Scholar] [CrossRef]
- Daikos, O.; Scherzer, T. In-line monitoring of the residual moisture in impregnated black textile fabrics by hyperspectral imaging. Prog. Org. Coat. 2021, 163, 106610. [Google Scholar] [CrossRef]
- Wang, C.; Xu, M.; Jiang, Y.; Zhang, G.; Cui, H.; Deng, G.; Lu, Z. Toward Real Hyperspectral Image Stripe Removal via Direction Constraint Hierarchical Feature Cascade Networks. Remote Sens. 2022, 14, 467. [Google Scholar] [CrossRef]
- Blanch-Perez-del-Notario, C.; Saeys, W.; Lambrechts, A. Hyperspectral imaging for textile sorting in the visible–near infrared range. J. Spectr. Imaging 2019, 8, a17. [Google Scholar] [CrossRef]
- Mirschel, G.; Daikos, O.; Scherzer, T. In-line monitoring of the thickness distribution of adhesive layers in black textile laminates by hyperspectral imaging. Comput. Chem. Eng. 2019, 124, 317–325. [Google Scholar] [CrossRef]
- Feng, X.; Cheng, H.; Zuo, D.; Zhang, Y.; Wang, Q.; Lv, L.; Li, S.; Yu, J.Z.; Song, G. Genome-wide identification and expression analysis of GL2-interacting-repressor (GIR) genes during cotton fiber and fuzz development. Planta 2022, 255, 1–18. [Google Scholar] [CrossRef]
- Anthony, W.S. Improvement of the Marketability of Cotton Produced in Zones Affected by Stickiness; Gourlot, J.-P., Ed.; Common Fund for Commodities: Amsterdam, The Netherlands, 2001; p. 99. [Google Scholar]
- Rony, A.N.U. Technical Properties of Cotton Fiber, Textile Learner GmbH. Available online: https://textilelearner.net/technical-properties-of-cotton-fiber/ (accessed on 26 December 2020).
- Abidi, N.; Hequet, E.; Cabrales, L. Changes in sugar composition and cellulose content during the secondary cell wall biogenesis in cotton fibers. Cellulose 2010, 17, 153–160. [Google Scholar] [CrossRef]
- Calvo-Agudo, M.; Tooker, J.F.; Dicke, M.; Tena, A. Insecticide-contaminated honeydew: Risks for beneficial insects. Biol. Rev. 2021, 97, 664–678. [Google Scholar] [CrossRef]
- Balasubramanya, R.; Bhatawdekar, S.; Paralikar, K. A new method for reducing the stickiness of cotton. Text. Res. J. 1985, 55, 227–232. [Google Scholar] [CrossRef]
- Jumaniyazov, K.; Egamberdiev, F.; Abbazov, I. The Effect of Crop Type on Cotton Quality Indicators. Int. J. Adv. Res. Sci. Eng. Technol. 2020, 7, 13510–13518. [Google Scholar]
- Severino, L.; Leite, B.; Gambarra-Neto, F.; Araújo, J.; Medeiros, E. Detection and Quantification of Stickiness on Cotton Samples Using Near Infrared Hyperspectral Images Bremen Baumwollboerse. p. 8. Available online: https://baumwollboerse.de/kompetenzen/international-cotton-conference/vortraege/ (accessed on 6 December 2022).
- Gamble, G.R. Evaluation of cotton stickiness via the thermochemical production of volatile compounds. J. Cotton Sci. 2003, 7, 45–50. [Google Scholar]
- Jiang, Y.; Li, C. Detection and discrimination of cotton foreign matter using push-broom based hyperspectral imaging: System design and capability. PLoS ONE 2015, 10, e0121969. [Google Scholar] [CrossRef]
- Abidi, N.; Hequet, E. Fourier transform infrared analysis of cotton contamination. Text. Res. J. 2007, 77, 77–84. [Google Scholar] [CrossRef]
- Was-Gubala, J.; Starczak, R. Nondestructive identification of dye mixtures in polyester and cotton fibers using Raman spectroscopy and ultraviolet-visible (UV-Vis) microspectrophotometry. Appl. Spectrosc. 2015, 69, 296–303. [Google Scholar] [CrossRef]
- Fortier, C.A.; Rodgers, J.E.; Cintron, M.S.; Cui, X.; Foulk, J.A. Identification of cotton and cotton trash components by Fourier transform near-infrared spectroscopy. Text. Res. J. 2011, 81, 230–238. [Google Scholar] [CrossRef]
- Mustafic, A.; Jiang, Y.; Li, C. Cotton contamination detection and classification using hyperspectral fluorescence imaging. Text. Res. J. 2016, 86, 1574–1584. [Google Scholar] [CrossRef]
- Miller, W.B.; Peralta, E.; Ellis, D.R.; Perkins Jr, H.H. Stickiness potential of individual insect honeydew carbohydrates on cotton lint. Text. Res. J. 1994, 64, 344–350. [Google Scholar] [CrossRef]
- Ghule, A.V.; Chen, R.K.; Tzing, S.H.; Lo, J.; Ling, Y.C. Simple and rapid method for evaluating stickiness of cotton using thermogravimetric analysis. Anal. Chim. Acta 2004, 502, 251–256. [Google Scholar] [CrossRef]
- Barton, F.; Bargeron III, J.; Gamble, G.; McAlister, D.; Hequet, E. Analysis of sticky cotton by near-infrared spectroscopy. Appl. Spectrosc. 2005, 59, 1388–1392. [Google Scholar] [CrossRef]
- Tschannerl, J.; Ren, J.; Jack, F.; Krause, J.; Zhao, H.; Huang, W.; Marshall, S. Potential of UV and SWIR hyperspectral imaging for determination of levels of phenolic flavour compounds in peated barley malt. Food Chem. 2019, 270, 105–112. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]
- Mika, S.; Ratsch, G.; Weston, J.; Scholkopf, B.; Mullers, K.-R. Fisher discriminant analysis with kernels. In Proceedings of the Neural Networks for Signal Processing IX: Proceedings of the 1999 IEEE Signal Processing Society Workshop (Cat. No. 98th8468), Madison, WI, USA, 25 August 1999; pp. 41–48. [Google Scholar]
- Stefanakis, M.; Lorenz, A.; Bartsch, J.W.; Bassler, M.C.; Wagner, A.; Brecht, M.; Pagenstecher, A.; Schittenhelm, J.; Boldrini, B.; Hakelberg, S. Formalin Fixation as Tissue Preprocessing for Multimodal Optical Spectroscopy Using the Example of Human Brain Tumour Cross Sections. J. Spectrosc. 2021, 2021, 1–14. [Google Scholar] [CrossRef]
- van Kollenburg, G.; Bouman, R.; Offermans, T.; Gerretzen, J.; Buydens, L.; van Manen, H.-J.; Jansen, J. Process PLS: Incorporating substantive knowledge into the predictive modelling of multiblock, multistep, multidimensional and multicollinear process data. Comput. Chem. Eng. 2021, 154, 107466. [Google Scholar] [CrossRef]
- Fischer, M.K.; Völkl, W.; Hoffmann, K.H. Honeydew production and honeydew sugar composition of polyphagous black bean aphid, Aphis fabae (Hemiptera: Aphididae) on various host plants and implications for ant-attendance. Eur. J. Entomol. 2005, 102, 155–160. [Google Scholar] [CrossRef]
- Hogervorst, P.A.; Wäckers, F.L.; Romeis, J. Effects of honeydew sugar composition on the longevity of Aphidius ervi. Entomol. Exp. Et Appl. 2007, 122, 223–232. [Google Scholar] [CrossRef]
- Victorita, B.; Marghitas, L.A.; Stanciu, O.; Laslo, L.; Dezmirean, D.; Bobis, O. High-performance liquid chromatographic analysis of sugars in Transylvanian honeydew honey. Bull. UASVM Anim. Sci. Biotechnol. 2008, 65, 229–232. [Google Scholar]
- The Journey of Cotton: Purification, Barnhardt Natural Fibers. Available online: https://barnhardtcotton.net/blog/journey-cotton-purification/ (accessed on 1 February 2022).
- Shepard, C.L.; Louis, G.L.; Simpson, J. Processing Mechanically Cleaned and Shortened Scoured Wool on the Cotton System. Text. Res. J. 1983, 53, 706–711. [Google Scholar] [CrossRef]
- EN 14278-2:2004; Textiles-Determination of Cotton Fibre Stickiness-Part 2: Method Using an Automatic Thermodetection Plate Device. Beuth Verlag GmbH: Berlin, Germany, 2004; p. 11.
- Boldrini, B.; Kessler, W.; Rebner, K.; Kessler, R.W. Hyperspectral imaging: A review of best practice, performance and pitfalls for in-line and on-line applications. J. Near Infrared Spectrosc. 2012, 20, 483–508. [Google Scholar] [CrossRef]
- Schlapfer, D.R.; Kaiser, J.W.; Brazile, J.; Schaepman, M.E.; Itten, K.I. Calibration concept for potential optical aberrations of the APEX pushbroom imaging spectrometer. In Proceedings of the Sensors, Systems, and Next-Generation Satellites VII, Barcelona, Spain, 2 February 2004; pp. 221–231. [Google Scholar]
- Calvini, R.; Ulrici, A.; Amigo, J. Sparse-Based Modeling of Hyperspectral Data. In Data Handling in Science and Technology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 30, pp. 613–634. [Google Scholar]
- Lottspeich, F.; Zorbas, H. Bioanalytik, 4th ed.; Spektrum, Akad. Verlag: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
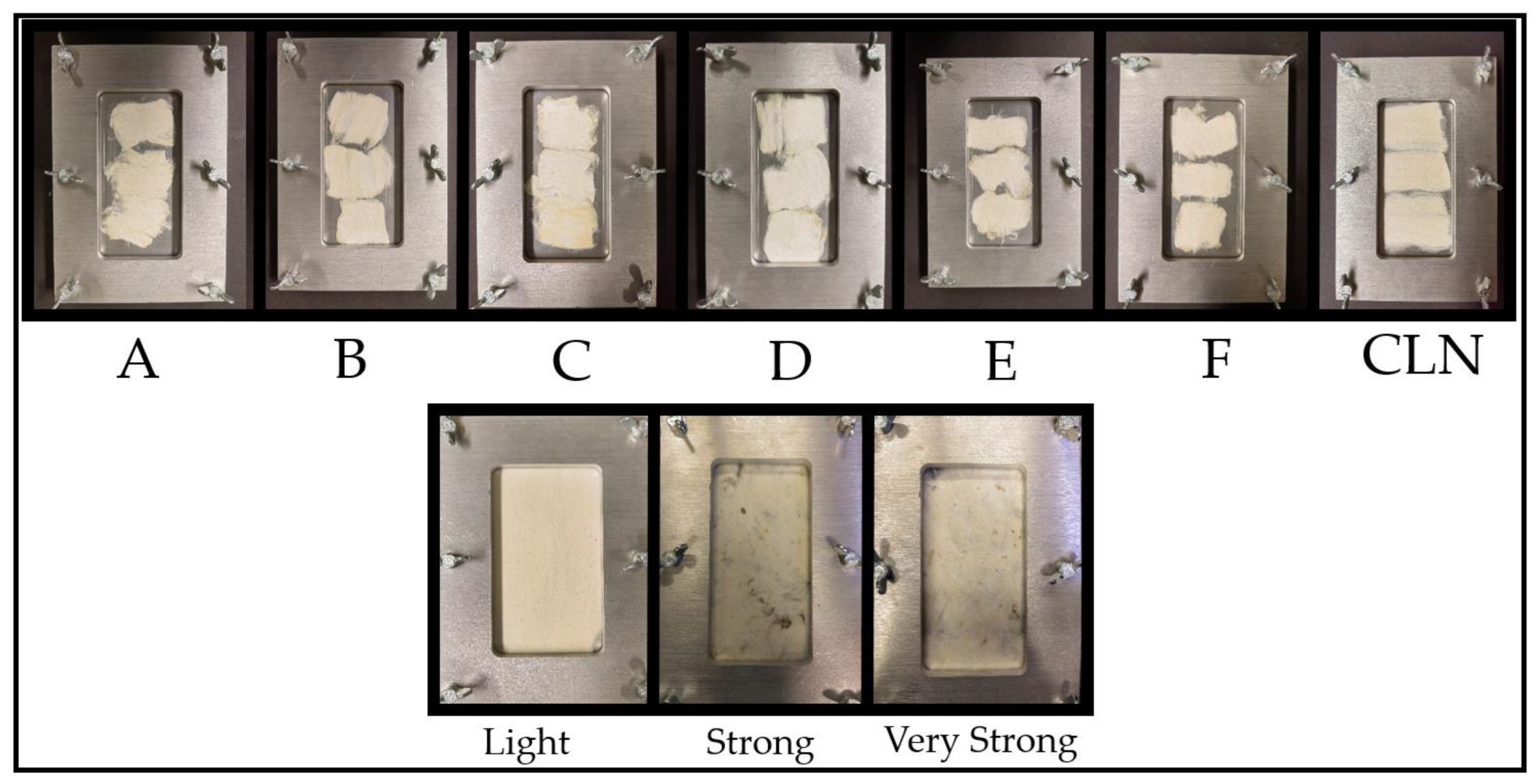


| Macronutrients and Natural Materials | Samples | Description | Manufacture | CAS Number |
|---|---|---|---|---|
| 1 | Glucose | D-Glucose anhydrous Laboratory reagent grade | Fisher Scientific GmbH, Leics, UK | 50-99-7 |
| 2 | Fructose | D-Fructose, 99.0% | ThermoFisher GmbH, Kandel, Germany | 57-48-7 |
| 3 | Sucrose | D-Sucrose, ≥99.9% For Molecular Biology | Fisher Scientific GmbH, Fair Lawn, NJ, USA | 57-50-1 |
| 4 | Melezitose | D-(+)-Melezitose monohydrate, ≥99.0% | Sigma-Aldric Chemie GmbH, Steinheim, Germany | 10030-67-8 |
| 5 | Trehalose | D- Trehalose anhydrous, 99.0% | Acros Organics, Fair Lawn, NJ, USA | 99-20-7 |
| 6 | Protein | Bovine Serum Albumin (BSA) fraction V, lyophilized powder | PAN-Biotech GmbH, Aidenbach, Germany | 9048-46-8 |
| Sample Type | Sugar Concentration/wt% | Ratio of: Sugar/g Dried Cotton/g |
|---|---|---|
| A | 2 | 0.2593 |
| B | 1 | 0.1331 |
| C | 0.5 | 0.0743 |
| D | 0.25 | 0.0386 |
| E | 0.125 | 0.0326 |
| F | 0.0625 | 0.0322 |
| CLN | - | - |
| Stickiness Type | Single Measurements | Average Number of Sticky Points | Sample |
|---|---|---|---|
| Light | 2, 11, 5 | 6 | 4301 |
| Strong | 47, 45, 47 | 46 | Sudan Girba Acala 3SG |
| Very strong | 60, 69, 80 | 70 | Sudan Gezira Acala type 3SG |
| Actual | ||||||||
|---|---|---|---|---|---|---|---|---|
| Predicted | Samples | A | B | C | D | E | F | CLN |
| A | 818 | 33 | 1 | 0 | 1 | 1 | 0 | |
| B | 33 | 554 | 93 | 42 | 15 | 45 | 0 | |
| C | 10 | 148 | 405 | 72 | 0 | 33 | 0 | |
| D | 0 | 69 | 66 | 642 | 34 | 143 | 0 | |
| E | 0 | 6 | 0 | 36 | 664 | 290 | 0 | |
| F | 3 | 54 | 11 | 72 | 149 | 352 | 5 | |
| CLN | 0 | 0 | 0 | 0 | 1 | 0 | 1819 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Ktash, M.; Stefanakis, M.; Wackenhut, F.; Jehle, V.; Ostertag, E.; Rebner, K.; Brecht, M. Prediction of Honeydew Contaminations on Cotton Samples by In-Line UV Hyperspectral Imaging. Sensors 2023, 23, 319. https://doi.org/10.3390/s23010319
Al Ktash M, Stefanakis M, Wackenhut F, Jehle V, Ostertag E, Rebner K, Brecht M. Prediction of Honeydew Contaminations on Cotton Samples by In-Line UV Hyperspectral Imaging. Sensors. 2023; 23(1):319. https://doi.org/10.3390/s23010319
Chicago/Turabian StyleAl Ktash, Mohammad, Mona Stefanakis, Frank Wackenhut, Volker Jehle, Edwin Ostertag, Karsten Rebner, and Marc Brecht. 2023. "Prediction of Honeydew Contaminations on Cotton Samples by In-Line UV Hyperspectral Imaging" Sensors 23, no. 1: 319. https://doi.org/10.3390/s23010319
APA StyleAl Ktash, M., Stefanakis, M., Wackenhut, F., Jehle, V., Ostertag, E., Rebner, K., & Brecht, M. (2023). Prediction of Honeydew Contaminations on Cotton Samples by In-Line UV Hyperspectral Imaging. Sensors, 23(1), 319. https://doi.org/10.3390/s23010319






