Low-Cost, Real-Time Polymerase Chain Reaction System with Integrated RNA Extraction
Abstract
1. Introduction
2. Materials and Methods
2.1. System Architecture
2.2. Gel Electrophoresis Analysis
2.3. Heat-Assisted Extraction Protocol
2.4. Reverse Transcription Conditions
2.5. PCR Conditions
2.6. Heat-Assisted RNA Extraction Efficiency Assessment
2.7. RT-PCR Performance Comparison between the Custom PCR and Commercial Systems
3. Results
3.1. Heat-Assisted Extraction Protocol
3.2. Heat-Assisted RNA Extraction Efficiency Assessment
3.3. RT-PCR Performance Comparison between Custom RT-PCR and Commercial Unit
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
- -
- Product Contents
- ⚬
- RNA Lysis Buffer
- ⚬
- RNA Prep Buffer
- ⚬
- RNA Wash Buffer (concentrate)
- ⚬
- DNase/RNase-Free Water
- ⚬
- DNase I2 (lyophilized)
- ⚬
- DNA Digestion Buffer
- ⚬
- Zymo-Spin™ IC Columns
- ⚬
- Collection Tubes
- ⚬
- Instruction Manual
- -
- Specifications
- ⚬
- Sample Sources—Cells (animal, gram(-) bacteria), soft and easy-to-lyse tissue, samples in DNA/RNA Shield™ or other preservation reagents, and enzymatic reactions (e.g., DNase I treated, Proteinase K treated). Not compatible with whole blood1 and urine1 samples.
- ⚬
- Size—Total RNA including small/microRNAs (≥17 nt).
- ⚬
- Purity—A260/A280 & A260/A230 > 1.8. RNA is ready for Next-Gen Sequencing, RT/qPCR, etc. Trace DNA can be removed by DNase I digestion.
- ⚬
- Binding Capacity—Zymo-Spin™ IC Column yield up to 10 µg RNA.
- ⚬
- Compatibility—For samples stored in preservation reagents: DNA/RNA Shield™, RNAprotect®, Allprotect®, Universal transport medium/viral transport medium (UTM®/VTM®), and RNAlater™.
- ⚬
- Elution Volume—≥6 µL DNase/RNase-Free Water.
- ⚬
- Equipment Needed (user provided)—Microcentrifuge, vortex.
- -
- Protocol
- ⚬
- Buffer Preparation
- ▪
- Add 96 mL of 100% ethanol to the 24 mL of RNA Wash Buffer concentrate (R1050).
- ▪
- Reconstitute lyophilized DNase I with DNase/RNase-Free Water, mix by gentle inversion, and store frozen aliquots: add 55 µL water.
- ⚬
- Sample Preparation: Perform all steps at room temperature and centrifugation at 10,000–16,000× g for 30 s, unless specified.
- ▪
- Remove cells liquid media from the culture container. Then add RNA Lysis Buffer directly to the monolayer. Remove cells from the culture surface by scraping, pipetting, etc.
- ⚬
- Total RNA Purification: Perform all steps at room temperature and centrifugation at 10,000–16,000× g for 30 s, unless specified.
- 1.
- Add 1 volume ethanol (95–100%) to 1 volume sample lysed in RNA Lysis Buffer (1:1) and mix well.
- 2.
- Transfer the mixture into a Zymo-Spin™ IC Column in a Collection Tube and centrifuge. Discard the flow-through.
- 3.
- DNase I treatment (recommended)
- Wash the column with 400 µL of RNA Wash Buffer and centrifuge. Discard the flowthrough.
- In a nuclease-free tube, add 5 µL of DNase I (1 U/µL) and 35 µL of DNA Digestion Buffer and mix. Add mixture directly into the column matrix.
- Incubate the column at room temperature (20–30 °C) for 15 min.
- 4.
- Add 400 µL of RNA Prep Buffer to the column and centrifuge. Discard the flow-through.
- 5.
- Add 700 µL of RNA Wash Buffer to the column and centrifuge. Discard the flow-through.
- 6.
- Add 400 µL of RNA Wash Buffer and centrifuge the column for 1 min to ensure complete removal of the wash buffer. Then carefully, transfer the column into a nuclease-free tube (not provided).
- 7.
- Add 15 µL of DNase/RNase-Free Water directly to the column matrix and centrifuge.
References
- World Health Organization. Who Coronavirus (COVID-19) Dashboard. World Health Organization. Available online: https://covid19.who.int/ (accessed on 15 January 2022).
- Pritt, B.S.; Wang, P.; Nuzzo, J.; Zimmermann, S.; Burnham, C.-A.D. Deadly pathogens, transformative technologies, and protracted pandemics: Challenges and opportunities in laboratory medicine. Clin. Chem. 2021, 68, 1–3. [Google Scholar] [CrossRef]
- Bong, C.-L.; Brasher, C.; Chikumba, E.; McDougall, R.; Mellin-Olsen, J.; Enright, A. The COVID-19 pandemic: Effects on low- and middle-income countries. Anesth. Analg. 2020, 131, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhang, H.; Xu, Y.; Laššáková, S.; Korabečná, M.; Neužil, P. PCR past, present and future. BioTechniques 2020, 69, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Garibyan, L.; Avashia, N. Polymerase chain reaction. J. Investig. Dermatol. 2013, 133, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Wright, W.F.; Simner, P.J.; Carroll, K.C.; Auwaerter, P.G. Progress report: Next-generation sequencing (NGS), multiplex polymerase chain reaction (PCR), and broad-range molecular assays as diagnostic tools for fever of unknown origin (FUO) investigations in adults. Clin. Infect. Dis. 2021, 74, 924–932. [Google Scholar] [CrossRef] [PubMed]
- Preston, C.M.; Harris, A.; Ryan, J.P.; Roman, B.; Marin, R., III; Jensen, S.; Everlove, C.; Birch, J.; Dzenitis, J.M.; Pargett, D.; et al. Underwater Application of quantitative PCR on an ocean mooring. PLoS ONE 2011, 6, e22522. [Google Scholar] [CrossRef]
- Liu, P.; Seo, T.S.; Beyor, N.; Shin, K.-J.; Scherer, J.R.; Mathies, R.A. Integrated portable polymerase chain reaction-capillary electrophoresis microsystem for rapid forensic short tandem repeat typing. Anal. Chem. 2007, 79, 1881–1889. [Google Scholar] [CrossRef]
- Noviyanti, F.; Shimizu, S.; Hosotani, Y.; Koseki, S.; Inatsu, Y.; Kawasaki, S. Predictive growth model of listeria monocytogenes under fluctuating temperature conditions in pasteurized milk by using real-time polymerase chain reaction. Foodborne Pathog. Dis. 2020, 17, 693–700. [Google Scholar] [CrossRef]
- Shafi, A.; Farooq, U.; Akram, K.; Khan, M.Z.; Hayat, Z.; Hayat, K. Molecular epidemiology of foodborne diseases. In Sequencing Technologies in Microbial Food Safety and Quality; CRC Press: Boca Raton, FL, USA, 2021; pp. 109–154. [Google Scholar] [CrossRef]
- Prevalence of Private Drinking Water Wells is Associated with Salmonellosis Incidence in Maryland, USA: An Ecological Analysis Using Foodborne Diseases Active Surveillance Network (FoodNet) Data (2007–2016). Available online: https://www.researchgate.net/publication/351506261_Prevalence_of_Private_Drinking_Water_Wells_is_Associated_with_Salmonellosis_Incidence_in_Maryland_USA_An_Ecological_Analysis_Using_Foodborne_Diseases_Active_Surveillance_Network_FoodNet_Data_2007-2016 (accessed on 24 January 2022).
- Chan, K.; Wong, P.-Y.; Yu, P.; Hardick, J.; Wong, K.-Y.; Wilson, S.A.; Wu, T.; Hui, Z.; Gaydos, C.; Wong, S.S. A rapid and low-cost PCR thermal cycler for infectious disease diagnostics. PLoS ONE 2016, 11, e0149150. [Google Scholar] [CrossRef]
- A Micromachined Low-Power-Consumption Portable PCR System. Available online: https://www.researchgate.net/publication/267721096_A_Micromachined_Low-power-consumption_Portable_PCR_System (accessed on 15 January 2022).
- Schneegaß, I.; Bräutigam, R.; Köhler, J.M. Miniaturized flow-through PCR with different template types in a silicon chip thermocycler. Lab Chip 2001, 1, 42–49. [Google Scholar] [CrossRef]
- Hennig, M.; Braun, D. Convective polymerase chain reaction around Micro Immersion Heater. Appl. Phys. Lett. 2005, 87, 183901. [Google Scholar] [CrossRef]
- Oda, R.P.; Strausbauch, M.A.; Huhmer, A.F.; Borson, N.; Jurrens, S.R.; Craighead, J.; Wettstein, P.J.; Eckloff, B.; Kline, B.; Landers, J.P. Infrared-mediated thermocycling for ultrafast polymerase chain reaction amplification of DNA. Anal. Chem. 1998, 70, 4361–4368. [Google Scholar] [CrossRef]
- Kadja, T.; Liu, C.; Sun, Y.; Chodavarapu, V.P. Low-cost, real-time polymerase chain reaction system for point-of-care medical diagnosis. Sensors 2022, 22, 2320. [Google Scholar] [CrossRef] [PubMed]
- Center for Food Safety and Applied Nutrition. Listeria (listeriosis). U.S. Food and Drug Administration. Available online: https://www.fda.gov/food/foodborne-pathogens/listeria-listeriosis#:~:text=Listeria%20monocytogenes%20(L.,and%20other%20food%20preservation%20measures (accessed on 15 January 2022).
- Komiazyk, M.; Walory, J.; Kozinska, A.; Wasko, I.; Baraniak, A. Impact of the nucleic acid extraction method and the RT-qpcr assay on SARS-COV-2 detection in low-viral samples. Diagnostics 2021, 11, 2247. [Google Scholar] [CrossRef] [PubMed]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Available online: https://www.frontiersin.org/articles/10.3389/fmicb.2017.00108/full (accessed on 9 December 2022).
- Lõoke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of genomic DNA from yeasts for PCR-based applications. BioTechniques 2017, 62, 5. [Google Scholar] [CrossRef]
- de Almeida, I.N.; da Silva Carvalho, W.; Rossetti, M.L.; Costa, E.R.; de Miranda, S.S. Evaluation of six different DNA extraction methods for detection of mycobacterium tuberculosis by means of PCR-IS6110: Preliminary study. BMC Res. Notes 2013, 6, 1–6. [Google Scholar] [CrossRef]
- Zarzoso-Lacoste, D.; Corse, E.; Vidal, E. Improving pcr detection of prey in molecular diet studies: Importance of group-specific primer set selection and Extraction Protocol performances. Mol. Ecol. Resour. 2012, 13, 117–127. [Google Scholar] [CrossRef]
- Heller, L.C.; Davis, C.R.; Peak, K.K.; Wingfield, D.; Cannons, A.C.; Amuso, P.T.; Cattani, J. Comparison of methods for DNA isolation from food samples for detection of shiga toxin-producing Escherichia coli by real-time PCR. Appl. Environ. Microbiol. 2003, 69, 1844–1846. [Google Scholar] [CrossRef]
- Tell, L.A.; Foley, J.; Needham, M.L.; Walker, R.L. Comparison of four rapid DNA extraction techniques for conventional polymerase chain reaction testing of three mycobacterium spp. that affect birds. Avian Dis. 2003, 47, 1486–1490. [Google Scholar] [CrossRef]
- Elizaquível, P.; Aznar, R. Comparison of four commercial DNA extraction kits for PCR detection of listeria monocytogenes, salmonella, Escherichia coli O157:H7, and Staphylococcus aureus in fresh, minimally processed vegetables. J. Food Prot. 2008, 71, 2110–2114. [Google Scholar] [CrossRef]
- Wang, T.Y.; Wang, L.; Zhang, J.H.; Dong, W.H. A simplified universal genomic DNA extraction protocol suitable for PCR. Genet. Mol. Res. 2011, 10, 519–525. [Google Scholar] [CrossRef]
- Barza, R.; Patel, P.; Sabatini, L.; Singh, K. Use of a simplified sample processing step without RNA extraction for direct SARS-COV-2 RT-PCR detection. J. Clin. Virol. 2020, 132, 104587. [Google Scholar] [CrossRef]
- Pastorino, B.; Bessaud, M.; Grandadam, M.; Murri, S.; Tolou, H.J.; Peyrefitte, C.N. Development of a taqman® RT-PCR assay without RNA extraction step for the detection and quantification of African chikungunya viruses. J. Virol. Methods 2005, 124, 65–71. [Google Scholar] [CrossRef]
- Camacho-Sanchez, M.; Burraco, P.; Gomez-Mestre, I.; Leonard, J.A. Preservation of RNA and DNA from mammal samples under field conditions. Mol. Ecol. Resour. 2013, 13, 663–673. [Google Scholar] [CrossRef]
- Chibo, D.; Birch, C. Analysis of human coronavirus 229e spike and nucleoprotein genes demonstrates genetic drift between chronologically distinct strains. J. Gen. Virol. 2006, 87, 1203–1208. [Google Scholar] [CrossRef]
- Reverse Transcription Setup: Thermo Fisher Scientific—US. Available online: https://www.thermofisher.com/us/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/rt-education/reverse-transcription-setup.html (accessed on 15 December 2022).
- King, A. In Virus Taxonomy: Classification and Nomenclature of Viruses; Elsevier: Amsterdam, The Netherlands, 2012; pp. 806–828. [Google Scholar]
- Fenwick, C.; Croxatto, A.; Coste, A.T.; Pojer, F.; André, C.; Pellaton, C.; Farina, A.; Campos, J.; Hacker, D.; Lau, K.; et al. Changes in SARS-COV-2 spike versus nucleoprotein antibody responses impact the estimates of infections in population-based seroprevalence studies. J. Virol. 2021, 95, e01828-20. [Google Scholar] [CrossRef]
- Quick-RNA Microprep Kit—ZYMO Research. Available online: https://files.zymoresearch.com/protocols/_r1050_r1051_quick-rna_microprep_kit.pdf (accessed on 26 April 2023).
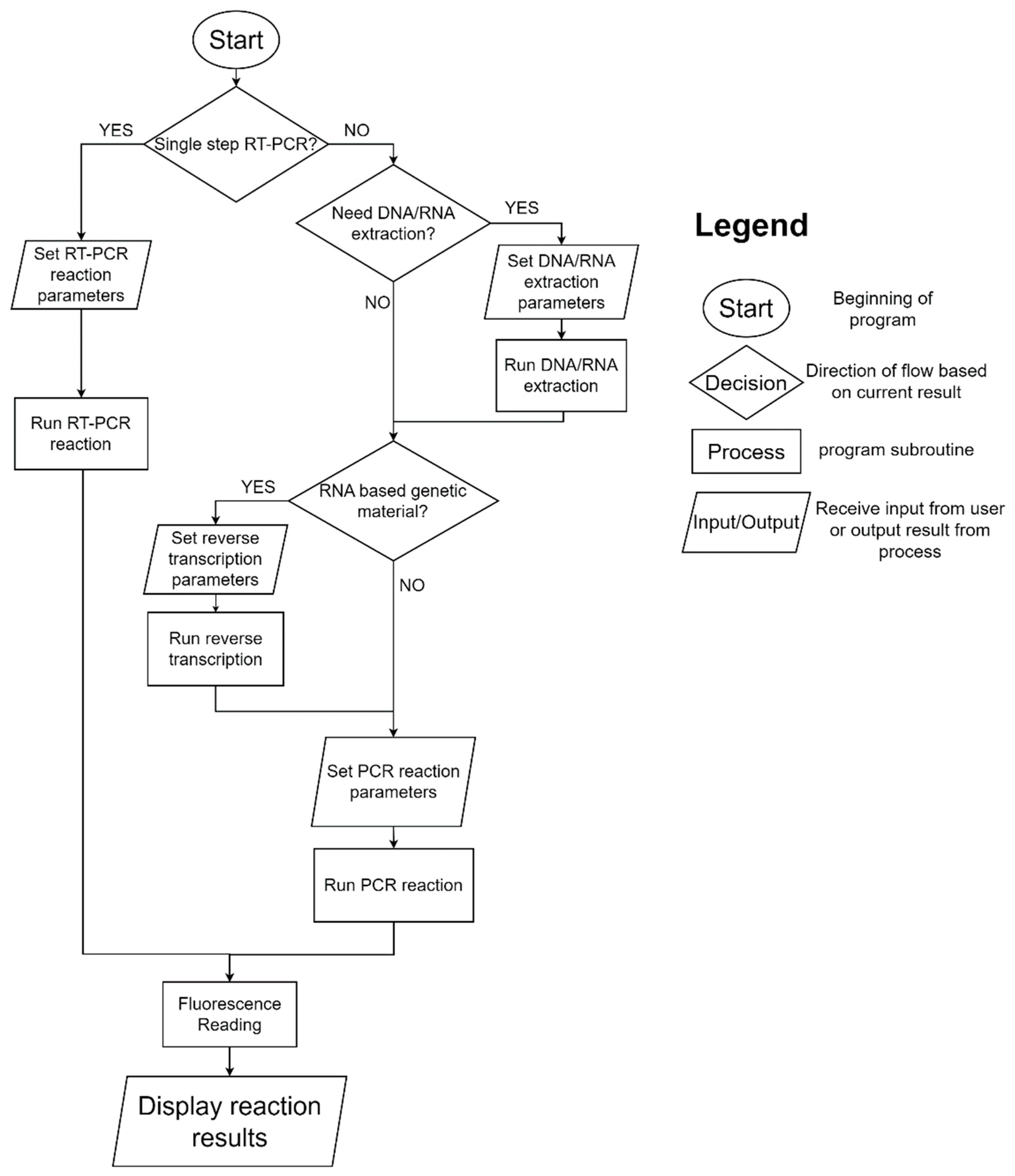
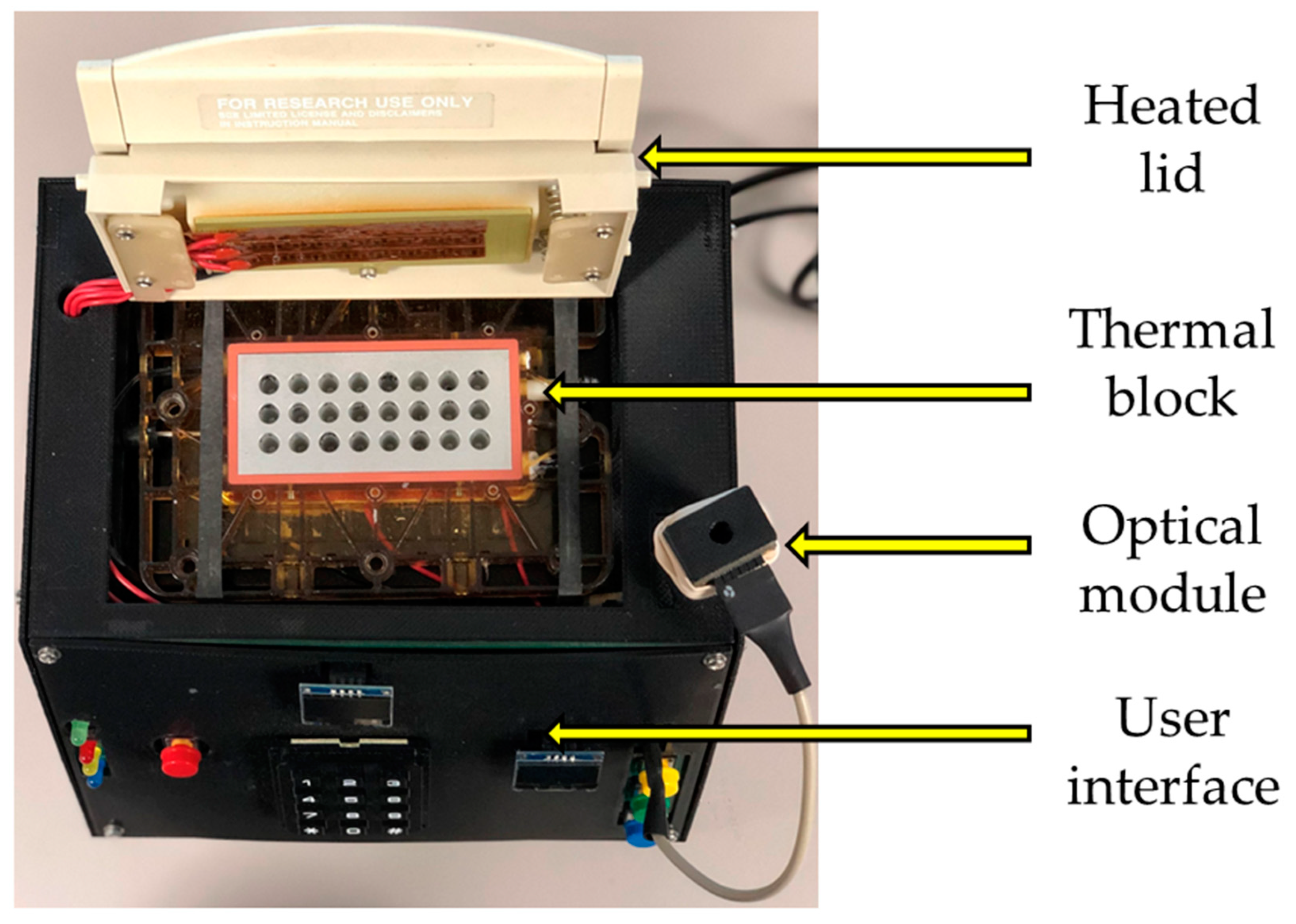

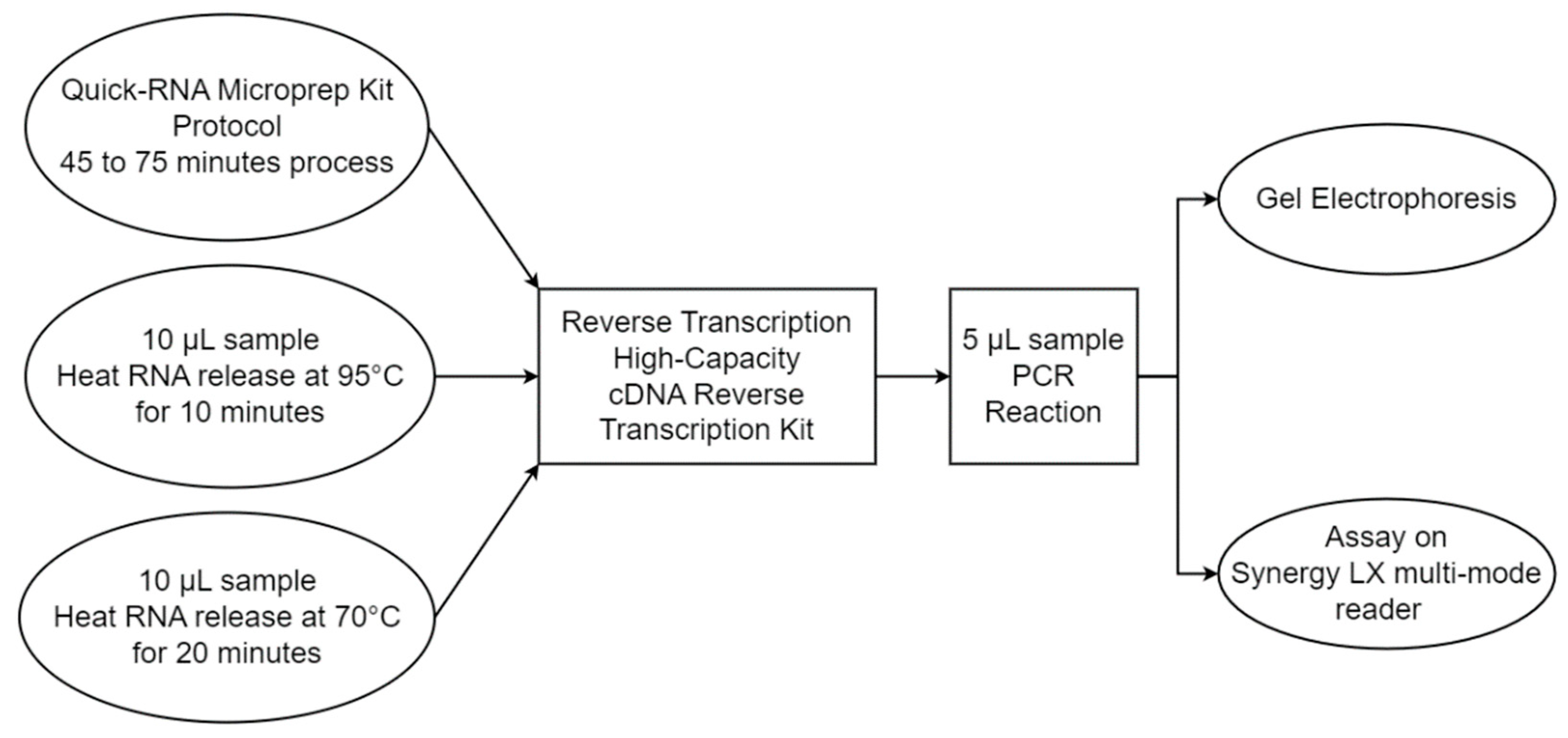
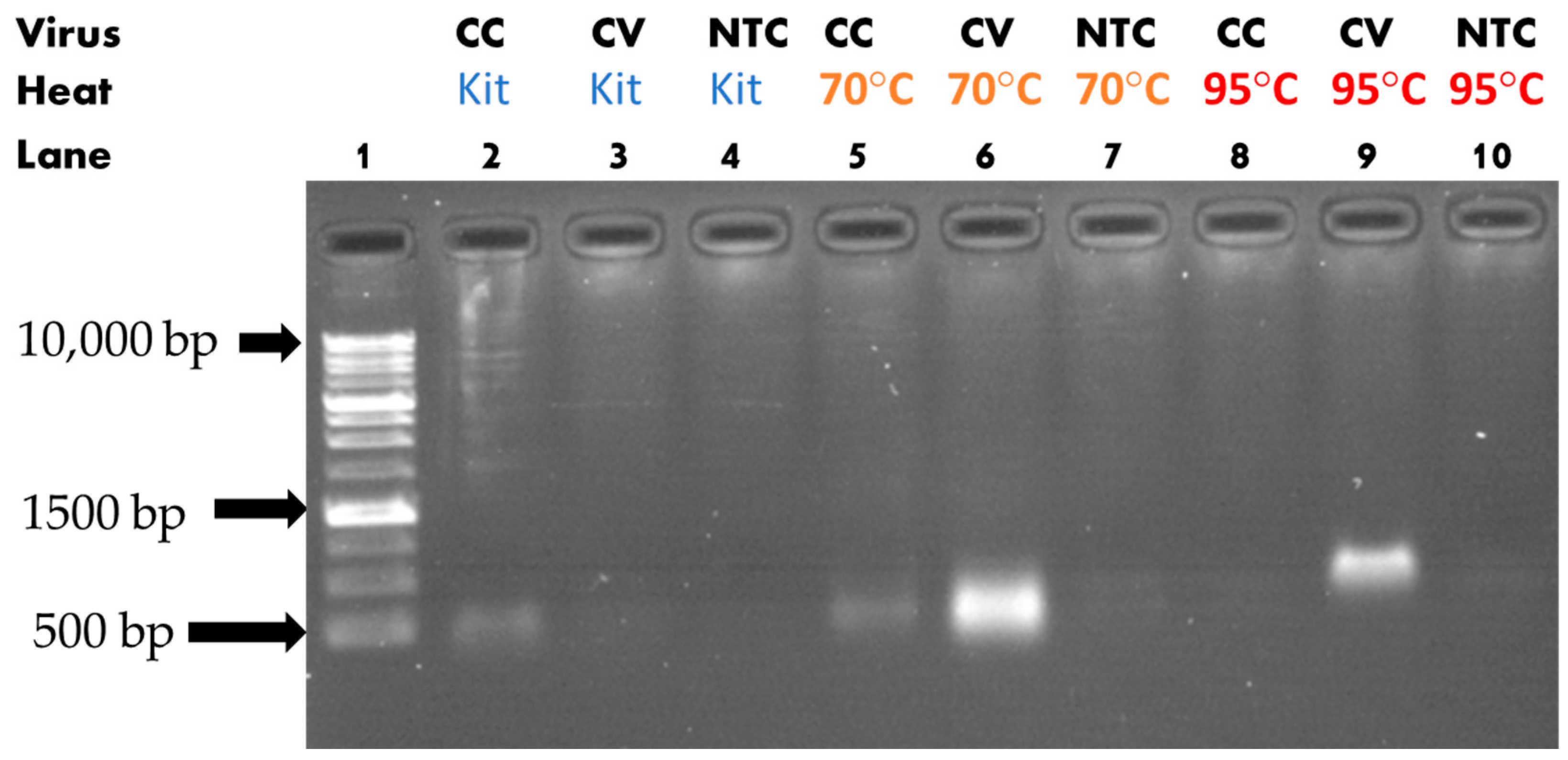
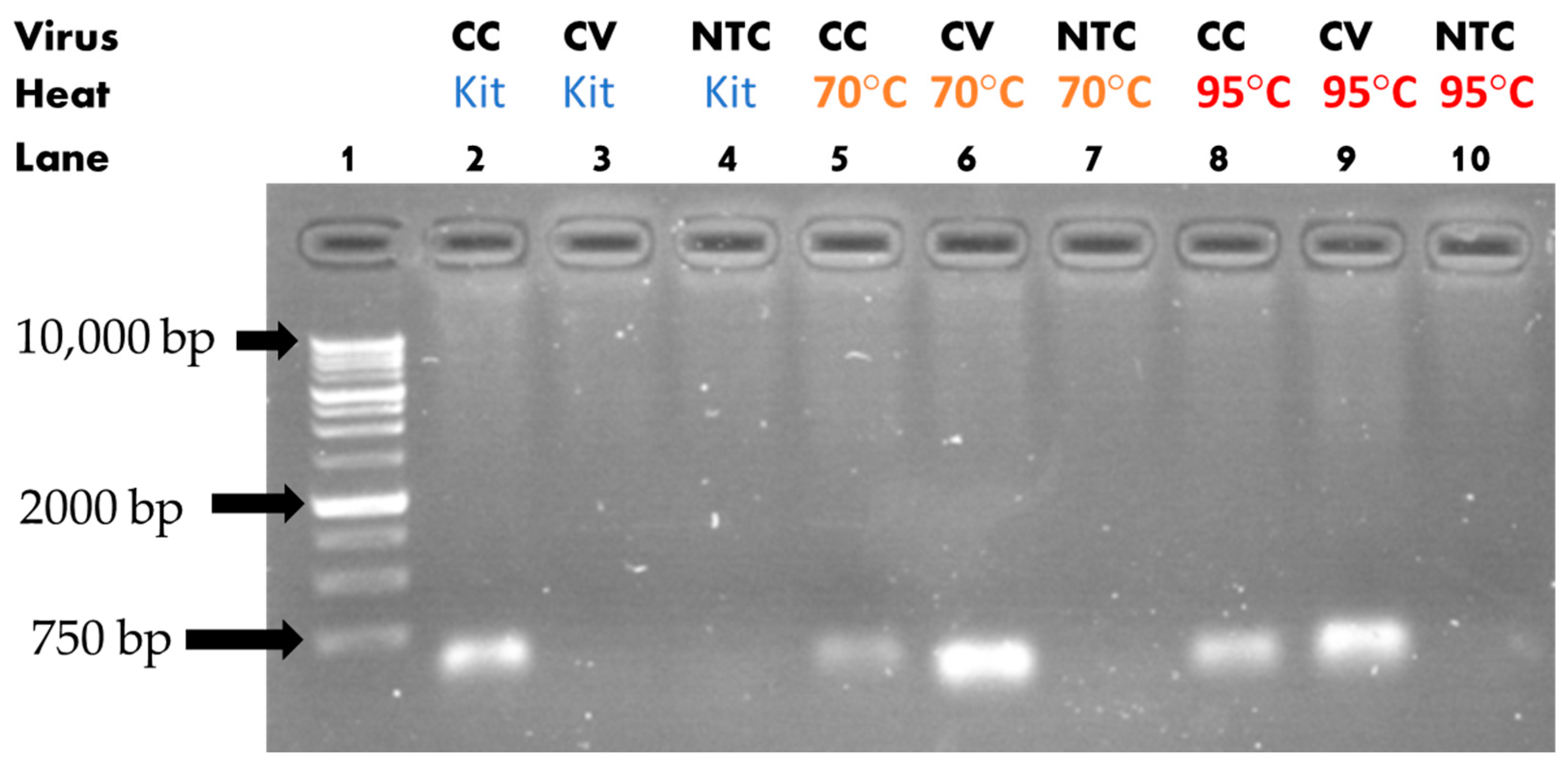
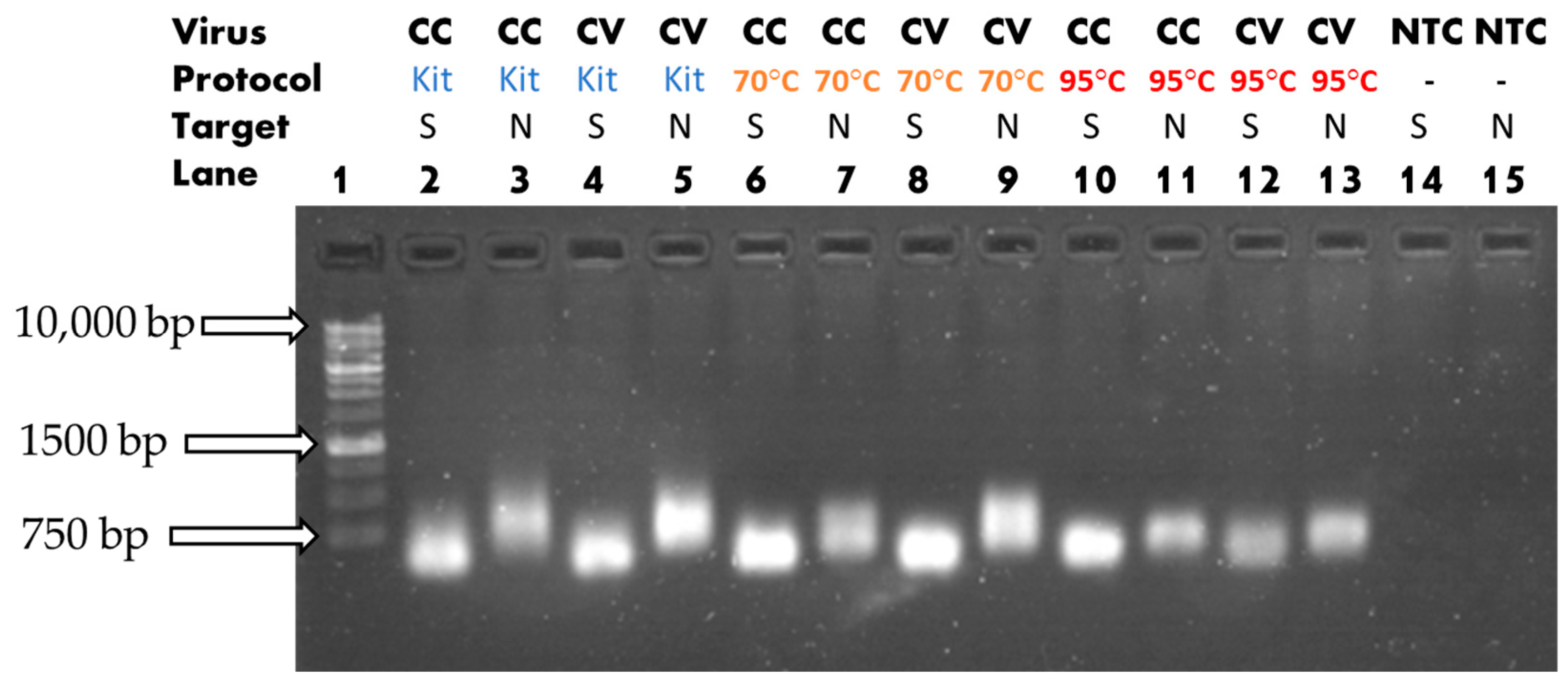
| Solution Type | Target Gene | ExtractionParameters | Reverse Transcription Reagents | PCR Reagents |
|---|---|---|---|---|
| Cell Culture/ Cultured Viral Solution/ dH2O (NTC) | Nucleoprotein (601 bp) | 70 °C/ 20 min 10 µL or 95 °C/ 10 min 10 µL or No heat 10 µL |
| 10 µL of Select Master Mix 1 µL of F. primer 5′- CTTGGAAGGTGATACCTCGTAAT -3′ 1 µL of R. primer 5′- GAACTCTGAGAACGAGCAAGA -3′ 3 µL of template (or 3 µL Nuclease-free H2O for NTC) 5 µL of deionized H2O |
| Cell Culture/ Cultured Viral Solution/ dH2O (NTC) | Spike (601 bp) | 70 °C/ 20 min 10 µL or 95 °C/ 10 min 10 µL or No heat 10 µL |
| 10 µL of Select Master Mix 1 µL of F. primer 5′- CAAGCTGTTGTTGGTGCTATG -3′ 1 µL of R. primer 5′- CAGTGCCAAGTCCAGAAGTAA -3′ 3 µL of template (or 3 µL Nuclease-free H2O for NTC) 5 µL of deionized H2O |
| Sample Type | Extraction Type | Before Extraction | After Extraction | A260/A280 (After Extraction) | Extraction Process Time | Extraction Cost |
|---|---|---|---|---|---|---|
| CC | Microprep Kit | 32.9 ng/µL (SD = 4.96) | 4.53 ng/µL (SD = 0.73) | 1.88 (SD = 0.54) | 45 to 75 min | $7 per reaction |
| 70 °C/20 min | 38.7 ng/µL (SD = 6.38) | 40.1 ng/µL (SD = 2.61) | 0.84 (SD = 0.04) | 20 min | ≈$0 | |
| 95 °C/10 min | 36.4 ng/µL (SD = 2.14) | 39.5 ng/µL (SD = 7.72) | 0.88 (SD = 0.04) | 10 min | ≈$0 | |
| CV | Microprep Kit | 144 ng/µL (SD = 32.4) | 10.8 ng/µL (SD = 10.7) | 2.40 (SD = 0.66) | 45 to 75 min | $7 per reaction |
| 70 °C/20 min | 144 ng/µL (SD = 32.4) | 224 ng/µL (SD = 42.5) | 0.81 (SD = 0.01) | 20 min | ≈$0 | |
| 95 °C/10 min | 144 ng/µL (SD = 32.4) | 210 ng/µL (SD = 35.1) | 0.88 (SD = 0.02) | 10 min | ≈$0 |
| Sample | Primer Type | Extraction Method | CT Value | CT Value Difference |
|---|---|---|---|---|
| CC | Nucleoprotein | Microprep Kit | 33.13 (SD = 2.82) | 0 |
| 70 °C/20 min | 36.57 (SD = 0.55) | +3.44 | ||
| 95 °C/10 min | Undet * | Undet | ||
| Spike | Microprep Kit | 32.80 (SD = 1.02) | 0 | |
| 70 °C/20 min | 32.67 (SD = 1.35) | −0.13 | ||
| 95 °C/10 min | 32.76 (SD = 1.55) | −0.04 | ||
| CV | Nucleoprotein | Microprep Kit | 33.32 (SD = 0.45) | 0 |
| 70 °C/20 min | 35.95 (SD = 0.77) | +2.63 | ||
| 95 °C/10 min | 36.44 (SD = 0.89) | +3.12 | ||
| Spike | Microprep Kit | 32.29 (SD = 0.46) | 0 | |
| 70 °C/20 min | 30.69 (SD = 1.36) | −1.60 | ||
| 95 °C/10 min | 31.87 (SD = 0.97) | −0.42 | ||
| NTC | Nucleoprotein | N/A | Undet | Undet |
| Spike | N/A | Undet | Undet |
| Tube | Device Type | Fluorescence Values | Fluorescence Difference with NTC |
| CC Spike 95 °C | StepOnePlus | 21.2 (SD = 6.46) | −4.57 |
| Custom RT-PCR | 445 (SD = 121) | +3.56 | |
| CC Nucleo 95 °C | StepOnePlus | 15.0 (SD = 0.67) | +4.12 |
| Custom RT-PCR | 545 (SD = 60.9) | −1.44 | |
| CV Spike 95 °C | StepOnePlus | 22.0 (SD = 6.44) | −3.82 |
| Custom RT-PCR | 523 (SD = 35.6) | +81.8 | |
| CV Nucleo 95 °C | StepOnePlus | 18.0 (SD = 6.75) | +7.12 |
| Custom RT-PCR | 574 (SD = 16.7) | +27.7 | |
| CC Spike 70 °C | StepOnePlus | 20.3 (SD = 8.91) | −5.53 |
| Custom RT-PCR | 444 (SD = 123) | +2 | |
| CC Nucleo 70 °C | StepOnePlus | 11.4 (SD = 2.70) | +0.49 |
| Custom RT-PCR | 506 (SD = 35.9) | −39.9 | |
| CV Spike 70 °C | StepOnePlus | 17.1 (SD = 7.17) | −8.74 |
| Custom RT-PCR | 525 (SD = 51.3) | +83.8 | |
| CV Nucleo 70 °C | StepOnePlus | 14.7 (SD = 2.84) | +3.83 |
| Custom RT-PCR | 611 (SD = 79.0) | +64.2 | |
| NTC Spike | StepOnePlus | 25.8 (SD = 7.05) | 0 |
| Custom RT-PCR | 441 (SD = 79.3) | 0 | |
| NTC Nucleo | StepOnePlus | 10.9 (SD = 12.7) | 0 |
| Custom RT-PCR | 546 (SD = 42.1) | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadja, T.; Sun, Y.; Chodavarapu, V.P. Low-Cost, Real-Time Polymerase Chain Reaction System with Integrated RNA Extraction. Sensors 2023, 23, 4604. https://doi.org/10.3390/s23104604
Kadja T, Sun Y, Chodavarapu VP. Low-Cost, Real-Time Polymerase Chain Reaction System with Integrated RNA Extraction. Sensors. 2023; 23(10):4604. https://doi.org/10.3390/s23104604
Chicago/Turabian StyleKadja, Tchamie, Yvonne Sun, and Vamsy P. Chodavarapu. 2023. "Low-Cost, Real-Time Polymerase Chain Reaction System with Integrated RNA Extraction" Sensors 23, no. 10: 4604. https://doi.org/10.3390/s23104604
APA StyleKadja, T., Sun, Y., & Chodavarapu, V. P. (2023). Low-Cost, Real-Time Polymerase Chain Reaction System with Integrated RNA Extraction. Sensors, 23(10), 4604. https://doi.org/10.3390/s23104604







