Real-Time Deep Recognition of Standardized Liver Ultrasound Scan Locations
Abstract
:1. Introduction
- We propose a scanning location-guided US system for accurate liver diagnosis that emphasizes the relationship between probe posture and the liver to improve accuracy and reduce diagnostic variability. Our method is the first to incorporate US-based scan location recognition for liver diagnosis by emphasizing the probe-posture and liver relationship.
- We develop a hierarchical deep learning architecture for liver scan location classification in US images by leveraging organ (liver, kidney, gallbladder) and vessel segmentation information and introducing a probabilistic representation method to enhance prediction robustness.
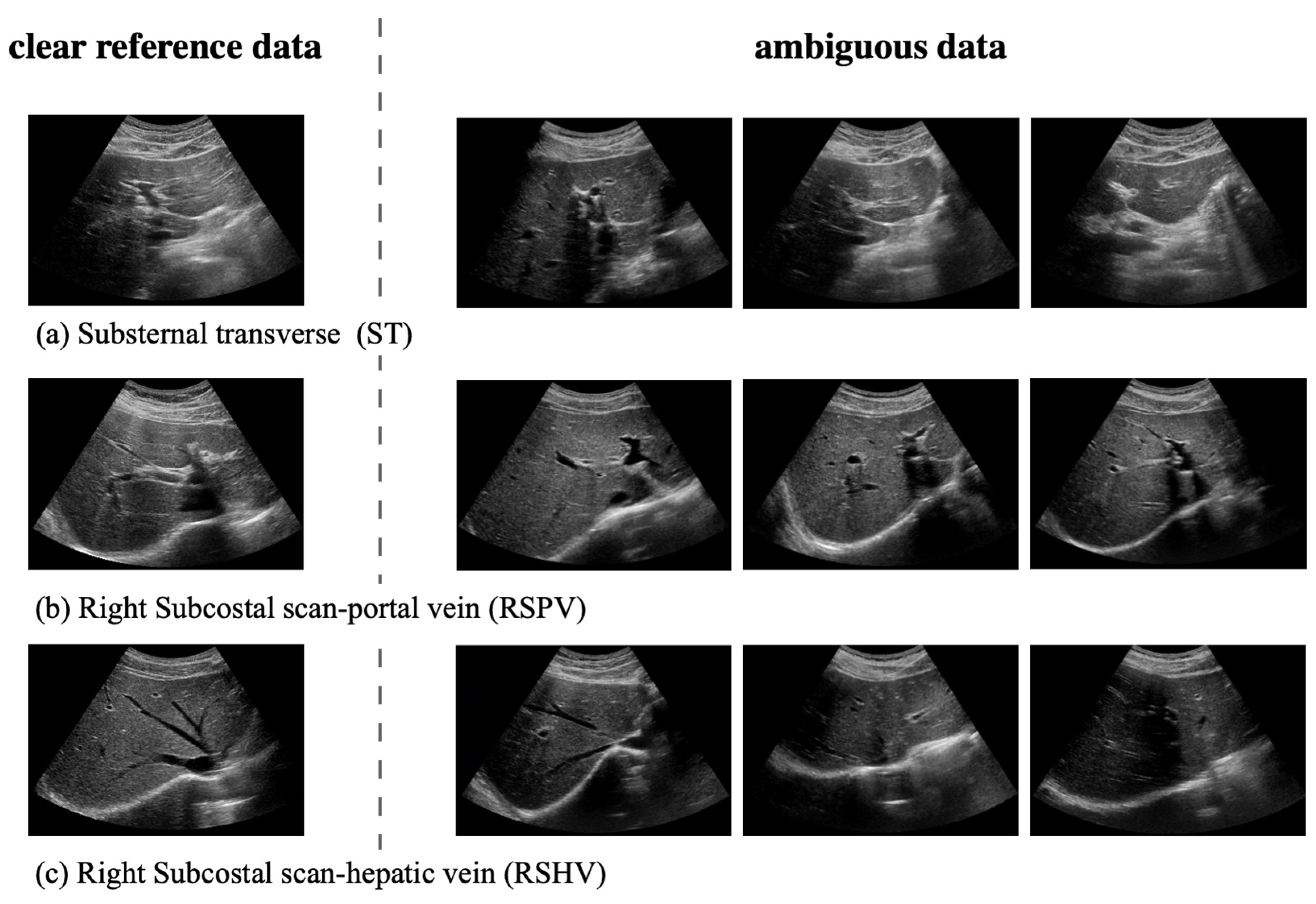
- We address the challenges of identifying key liver regions and handling ambiguous data, and we improve diagnostic accuracy in difficult cases by accounting for slight movement during US examinations.
2. Overview of the Proposed Approach
3. Datasets
4. Deep Hierarchical Network (DHN) for Standardized US Liver Scan (LS) Classification: LS-DHN
4.1. Feature Extraction from Segmentation Network
4.1.1. Global Long-Term Feature Extraction
4.1.2. Vessel-Related Feature Extraction
4.2. Architecture of the LS-DHN
4.2.1. Handling Ambiguous Data with Probabilistic Representation
4.2.2. First Hierarchy: Classification of Eight Liver Regions
4.2.3. Second Hierarchy: Hierarchical Classification of the ST, RSPV, and RSHV Classes by Handling Ambiguous Data
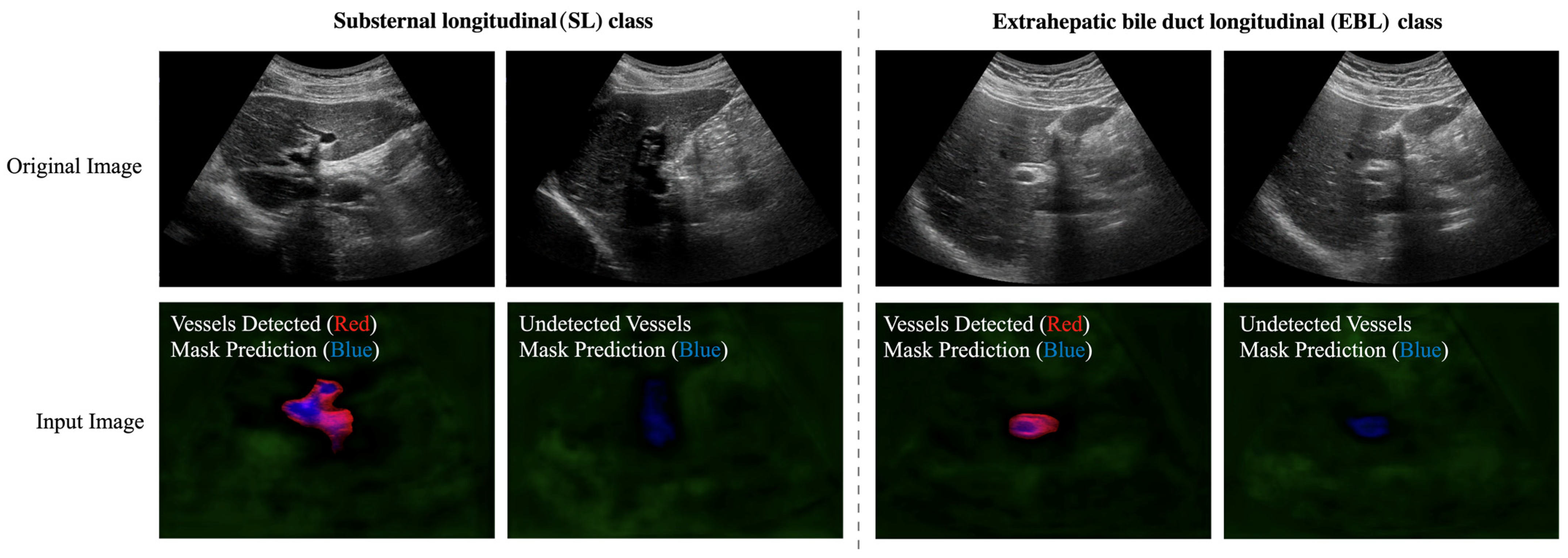
4.2.4. Second Hierarchy: Hierarchical Classification of the SL and EBL Classes
5. Results
5.1. Performance of Organ and Vessel Semantic Segmentation
5.2. Performance of MaskFormer-Based Liver US Scan Location Classification
5.2.1. Performance of the Hierarchical LS-DHN
5.2.2. Ablation Study: Non-Hierarchical Liver US Scan Location Classification
5.3. Discussion
6. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vernuccio, F.; Cannella, R.; Bartolotta, T.V.; Galia, M.; Tang, A.; Brancatelli, G. Advances in liver US, CT, and MRI: Moving toward the future. Eur. Radiol. Exp. 2021, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Das, T.K.; Chowdhary, C.L.; Gao, X.Z. Chest X-ray investigation: A convolutional neural network approach. J. Biomim. Biomater. Biomed. Eng. 2020, 45, 57–70. [Google Scholar] [CrossRef]
- Tragakis, A.; Kaul, C.; Murray-Smith, R.; Husmeier, D. The Fully Convolutional Transformer for Medical Image Segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 7 January 2023; pp. 3660–3669. [Google Scholar]
- Mishra, D.; Chaudhury, S.; Sarkar, M.; Soin, A.S. Ultrasound image segmentation: A deeply supervised network with attention to boundaries. IEEE Trans. Biomed. Eng. 2018, 66, 1637–1648. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Liu, S.; Yuan, D.; Wang, L.; Chen, J.; Lukasiewicz, T.; Fu, Z.; Zhang, R. ω-net: Dual supervised medical image segmentation with multi-dimensional self-attention and diversely-connected multi-scale convolution. Neurocomputing 2022, 500, 177–190. [Google Scholar] [CrossRef]
- Rhyou, S.-Y.; Yoo, J.-C. Cascaded deep learning neural network for automated liver steatosis diagnosis using ultrasound images. Sensors 2021, 21, 5304. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zeng, P.; Liu, P.; Lv, G. Automatic classification method of liver ultrasound standard plane images using pre-trained convolutional neural network. Connect. Sci. 2022, 34, 975–989. [Google Scholar] [CrossRef]
- Heinrich, M.P.; Siebert, H.; Graf, L.; Mischkewitz, S.; Hansen, L. Robust and Realtime Large Deformation Ultrasound Registration Using End-to-End Differentiable Displacement Optimisation. Sensors 2023, 23, 2876. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.-C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 6000–6010. [Google Scholar]
- Cheng, B.; Schwing, A.; Kirillov, A. Per-pixel classification is not all you need for semantic segmentation. Adv. Neural Inf. Process. Syst. 2021, 34, 17864–17875. [Google Scholar]
- Parnami, A.; Lee, M. Learning from few examples: A summary of approaches to few-shot learning. arXiv 2022, arXiv:2203.04291. [Google Scholar]
- Singh, R.; Bharti, V.; Purohit, V.; Kumar, A.; Singh, A.K.; Singh, S.K. MetaMed: Few-shot medical image classification using gradient-based meta-learning. Pattern Recognit. 2021, 120, 108111. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Huang, S.-W.; Lin, C.-T.; Chen, S.-P.; Wu, Y.-Y.; Hsu, P.-H.; Lai, S.-H. Auggan: Cross domain adaptation with gan-based data augmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 718–731. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- End, B.; Prats, M.I.; Minardi, J.; Sharon, M.; Bahner, D.P.; Boulger, C.T. Language of transducer manipulation 2.0: Continuing to codify terms for effective teaching. Ultrasound J. 2021, 13, 44. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]







| Model | Class | IoU | Pixel Acc |
|---|---|---|---|
| Organ | Liver | 85.57 | 92.44 |
| Kidney | 82.79 | 90.79 | |
| Gallbladder | 70.31 | 81.38 | |
| Vessel | 50.80 | 63.40 | |
| Method | w/Organ (Liver & Kidney & Gallbladder) Info | ||
|---|---|---|---|
| Class | Precision | Recall | F1-Score |
| ST, RSPV, RSHV | 0.97 | 0.99 | 0.98 |
| SL, EBL | 0.95 | 0.94 | 0.95 |
| RSLT | 0.94 | 0.86 | 0.90 |
| LD | 0.89 | 0.96 | 0.93 |
| GBL | 0.92 | 0.97 | 0.95 |
| RIA | 0.91 | 0.89 | 0.90 |
| RIP | 0.92 | 0.88 | 0.90 |
| LK | 1.00 | 0.99 | 0.99 |
| Method | (Before) Sub-Step 1: w/Liver & Vessel Info (after) Sub-Step 2: Probabilistic Representation w/Liver & Vessel Info | ||
|---|---|---|---|
| Class | Precision | Recall | F1-score |
| ST | 0.92 → 0.98 | 0.79 → 0.84 | 0.85 → 0.90 |
| RSPV | 0.88 → 0.95 | 0.90 → 0.98 | 0.89 → 0.96 |
| RSHV | 0.85 → 0.88 | 0.97 → 1.00 | 0.91 → 0.93 |
| Method | ImageNet Fine-Tuned ResNet-50 | Non-Hierarchical | LS-DHN | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Class | Precision | Recall | F1-score | Precision | Recall | F1-score | Precision | Recall | F1-score |
| SL | 0.56 | 0.81 | 0.66 | 0.74 | 0.88 | 0.81 | 0.88 | 0.98 | 0.93 |
| ST | 0.62 | 0.65 | 0.64 | 0.85 | 0.78 | 0.82 | 0.98 | 0.84 | 0.90 |
| RSPV | 0.73 | 0.61 | 0.66 | 0.91 | 0.85 | 0.88 | 0.95 | 0.98 | 0.96 |
| RSLT | 0.85 | 0.95 | 0.90 | 0.91 | 0.88 | 0.90 | 0.94 | 0.86 | 0.90 |
| LD | 0.88 | 0.84 | 0.86 | 0.91 | 0.97 | 0.94 | 0.89 | 0.96 | 0.93 |
| RSHV | 0.72 | 0.78 | 0.75 | 0.79 | 0.94 | 0.86 | 0.88 | 1.00 | 0.93 |
| GBL | 0.81 | 0.64 | 0.72 | 0.90 | 0.95 | 0.93 | 0.92 | 0.97 | 0.95 |
| EBL | 0.62 | 0.33 | 0.43 | 0.86 | 0.49 | 0.63 | 0.96 | 0.81 | 0.88 |
| RIA | 0.80 | 0.89 | 0.84 | 0.84 | 0.97 | 0.90 | 0.91 | 0.89 | 0.90 |
| RIP | 0.92 | 0.75 | 0.82 | 0.96 | 0.83 | 0.89 | 0.92 | 0.88 | 0.99 |
| LK | 0.85 | 0.89 | 0.87 | 1.00 | 0.99 | 1.00 | 1.00 | 0.99 | 0.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, J.; Lee, S.; Yi, J. Real-Time Deep Recognition of Standardized Liver Ultrasound Scan Locations. Sensors 2023, 23, 4850. https://doi.org/10.3390/s23104850
Shin J, Lee S, Yi J. Real-Time Deep Recognition of Standardized Liver Ultrasound Scan Locations. Sensors. 2023; 23(10):4850. https://doi.org/10.3390/s23104850
Chicago/Turabian StyleShin, Jonghwan, Sukhan Lee, and Juneho Yi. 2023. "Real-Time Deep Recognition of Standardized Liver Ultrasound Scan Locations" Sensors 23, no. 10: 4850. https://doi.org/10.3390/s23104850





