An End-to-End Multi-Channel Convolutional Bi-LSTM Network for Automatic Sleep Stage Detection
Abstract
:1. Introduction
- ▪
- This paper proposes a multi-channel, more specifically a four-channel, convolutional Bi-LSTM network for automatic sleep scoring with high accuracy.
- ▪
- In the proposed model, a dual-channel two-layer CNN-incorporated Bi-LSTM network module is designed and pre-trained utilizing data from any two distinct channel signals of the PSG recording.
- ▪
- Once the pre-training is finished and the dual channel module is validated, using two such pre-trained modules, a four-channel model has been developed, and the concept of transfer learning is employed circuitously to reduce the burden of a high computational cost and reduce the overall training time.
- ▪
- In the dual-channel module, convolutional layers are employed to extract spatial features from two channel PSG recordings, and these extracted spatial features are coupled with the input at every level of the BI-LSTM network to extract and learn rich temporally correlated features.
- ▪
- Finally, we have evaluated the performances of the proposed model on the Sleep EDF-20 and Sleep EDF-78 datasets. In addition, we compared the performance with other existing works.
2. Related Works
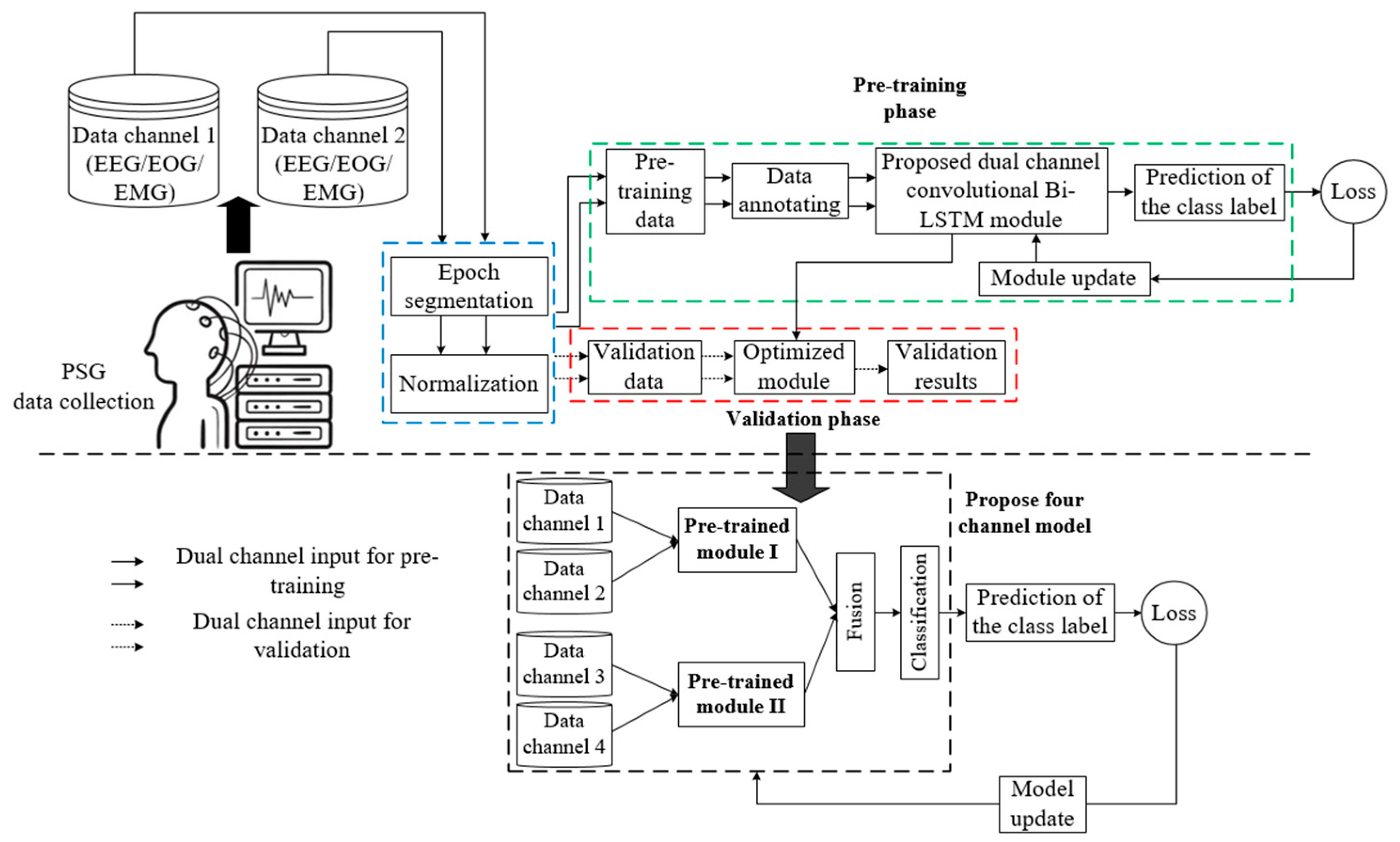
3. Materials and Methods
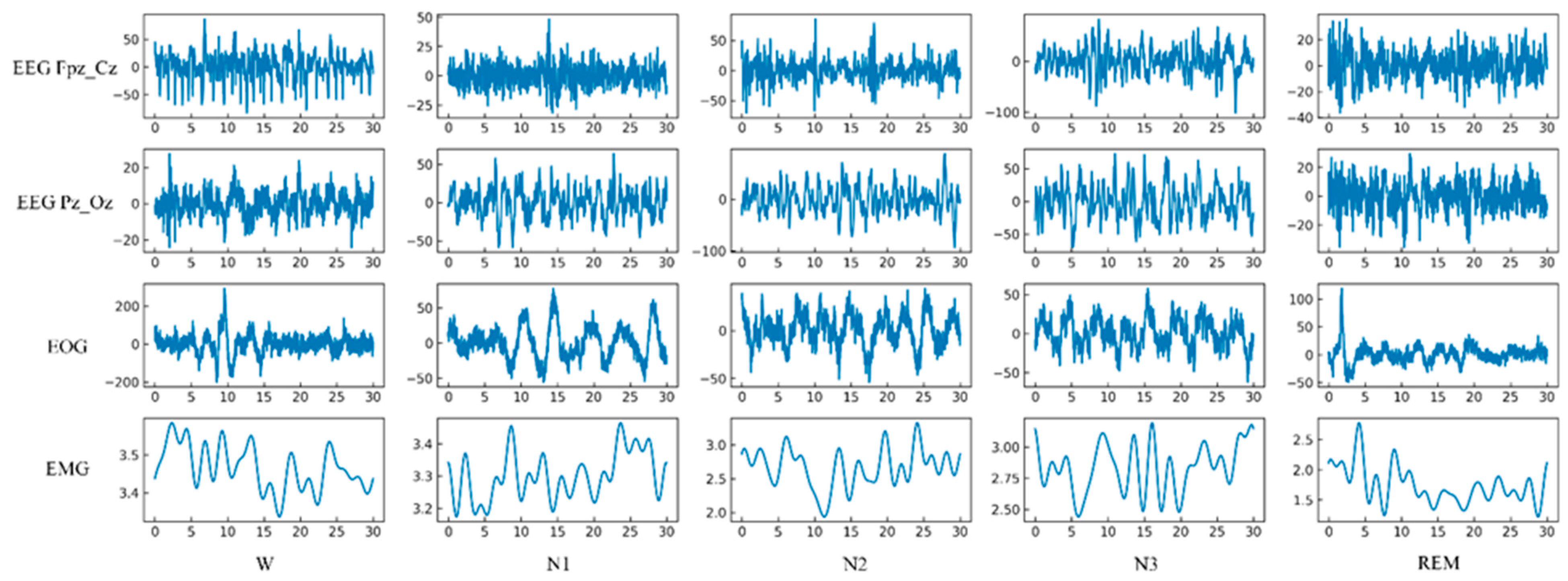
3.1. Sleep EDF Database Description
3.2. Epoch Segmentation and Data Annotating
3.3. Data Normalization and Splitting
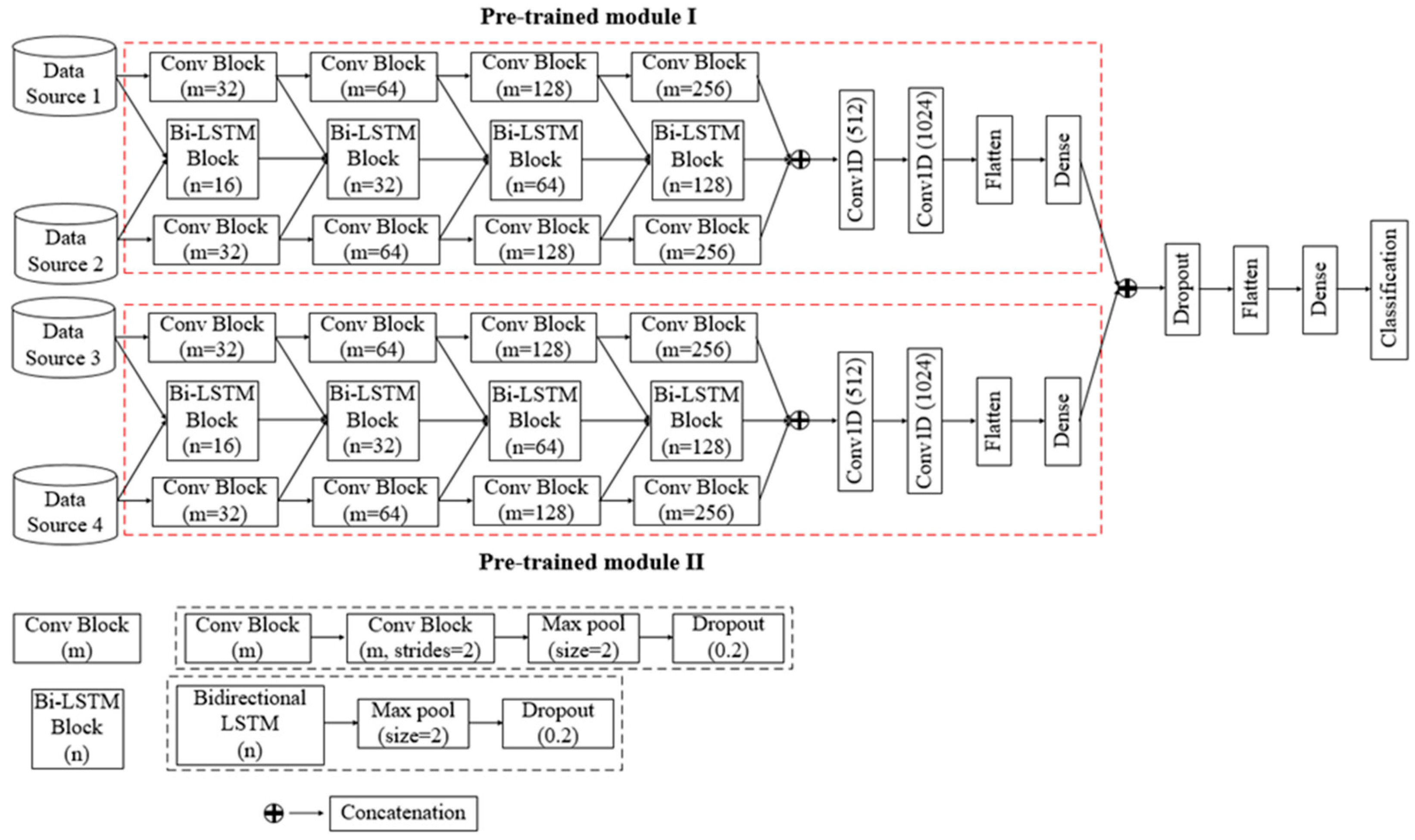
3.4. Four-Channel Convolutional Bi-LSTM Network
4. Experimental Results
4.1. Implementation Details and Performance Evaluation Metrics
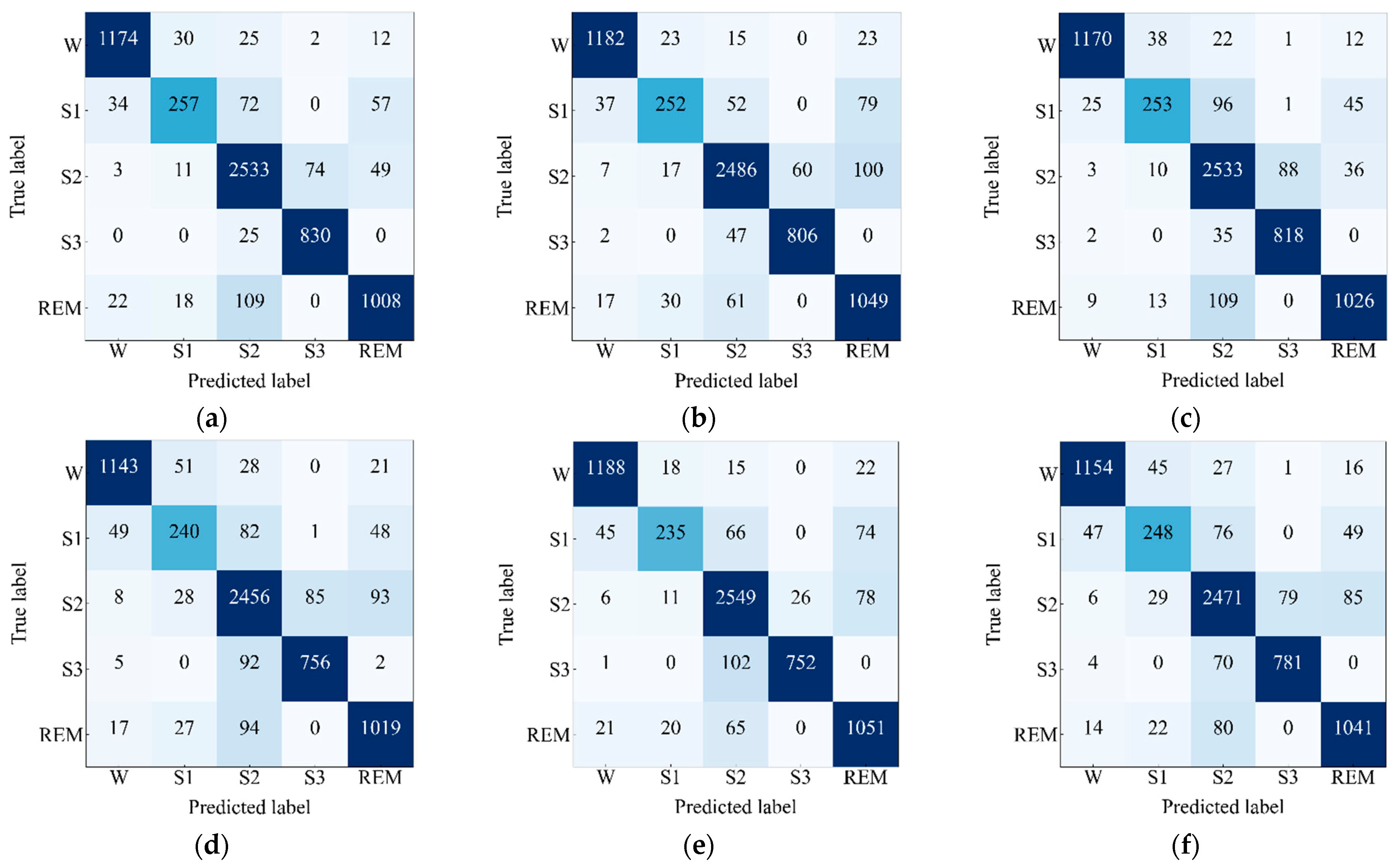
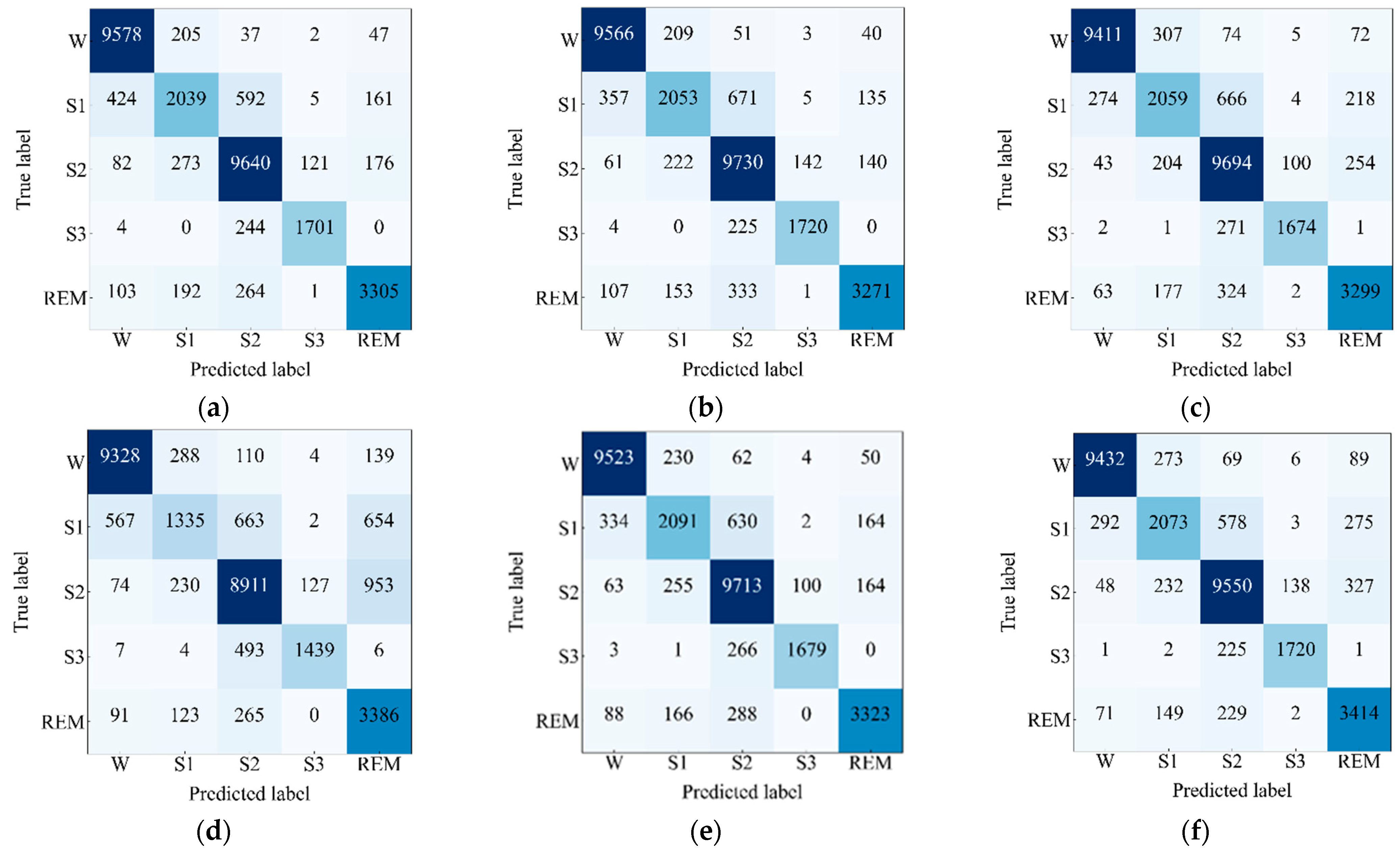
4.2. Performance Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wulff, K.; Gatti, S.; Wettstein, J.G.; Foster, R.G. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat. Rev. Neurosci. 2010, 11, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Redmond, S.J.; Heneghan, C. Cardiorespiratory-based sleep staging in subjects with obstructive sleep apnea. IEEE Trans. Biomed. Eng. 2006, 53, 485–496. [Google Scholar] [CrossRef]
- Iber, C.; Ancoli-Israel, S.; Chesson, A.; Quan, S.F. The AASM Manual for the Scoring of Sleep and Associated Events; American Academy of Sleep Medicine: Westchester, IL, USA, 2007. [Google Scholar]
- Rechtschaffen, A.; Kales, A. A Manual Standardized Terminology, Techniques, and Scoring System for Sleep Stages of Human Subjects; US Public Health Service, US Government Printing Office: Washington, DC, USA, 1968. [Google Scholar]
- Faisal, F.; Nishat, M.M.; Mahbub, M.A.; Shawon, M.M.I.; Alvi, M.M.-U.-H. COVID-19 and its impact on school closures: A predictive analysis using machine learning algorithms. In Proceedings of the 2021 International Conference on Science & Contemporary Technologies (ICSCT), Dhaka, Bangladesh, 5–7 August 2021; pp. 1–6. [Google Scholar]
- Lim, B.; Zohren, S. Time-series forecasting with deep learning: A survey. Philos. Trans. R. Soc. A 2021, 379, 20200209. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Toma, T.I.; Choi, S. A Parallel Cross Convolutional Recurrent Neural Network for Automatic Imbalanced ECG Arrhythmia Detection with Continuous Wavelet Transform. Sensors 2022, 22, 7396. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.H.; Zheng, X.W.; Shao, X.X.; Cui, L.Z. Automatic sleep stage classification based on convolutional neural network and fine-grained segments. Complexity 2018, 2018, 9248410. [Google Scholar] [CrossRef]
- Sors, A.; Bonnet, S.; Mirek, S.; Vercueil, L.; Payen, J.F. A convolutional neural network for sleep stage scoring from raw single-channel EEG. Biomed. Signal Process. 2018, 42, 107–114. [Google Scholar] [CrossRef]
- Supratak, A.; Dong, H.; Wu, C.; Guo, Y. DeepSleepNet: A Model for Automatic Sleep Stage Scoring Based on Single-Channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1998–2008. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.; Andreotti, F.; Cooray, N.; Chen, O.Y.; De Vos, M. Joint classification and prediction CNN framework for automatic sleep stage classification. IEEE Trans. Biomed. Eng. 2019, 66, 1285–1296. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Gao, Z.; An, J.; Gao, S.; Grebogi, C. A Graph-Temporal Fused Dual-Input Convolutional Neural Network for Detecting Sleep Stages from EEG Signals. IEEE Trans. Circuits Syst. II Express Briefs 2021, 68, 777–781. [Google Scholar] [CrossRef]
- Phan, H.; Chen, O.Y.; Koch, P.; Lu, Z.; McLoughlin, I.; Mertins, A. Towards More Accurate Automatic Sleep Staging via Deep Transfer Learning. IEEE Trans. Biomed. Eng. 2021, 68, 1787–1798. [Google Scholar] [CrossRef]
- Phan, H.; Chen, O.Y.; Tran, M.C.; Koch, P.; Mertins, A.; Vos, M.D. XSleepNet. Multi-view sequential model for automatic sleep staging. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 5903–5915. [Google Scholar] [CrossRef]
- Guillot, A.; Thorey, V. RobustSleepNet. Transfer Learning for Automated Sleep Staging at Scale. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- Tianqi, Z.; Wei, L.; Feng, Y. Multi-branch convolutional neural network for automatic sleep stage classification with embedded stage refinement and residual attention channel fusion. Sensors 2020, 20, 6592. [Google Scholar]
- Khalili, E.; Asl, B.M. Automatic sleep stage classification using temporal convolutional neural network and new data augmentation technique from raw single-channel EEG. Comput. Meth. Programs Biomed. 2021, 204, 106063. [Google Scholar] [CrossRef]
- Eldele, E.; Chen, Z.; Liu, C.; Wu, M.; Kwoh, C.K.; Li, X.; Guan, C. An attention-based deep learning approach for sleep stage classification with single-channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 809–818. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, M.; Li, Y.; Su, M.; Xu, Z.; Wang, C.; Kang, D.; Li, H.; Mu, X.; Ding, X.; et al. Automated multi-model deep neural network for sleep stage scoring with unfiltered clinical data. Sleep Breath. 2020, 24, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Ran, F.; Xu, M.; Guez, A.; Li, A.; Guo, A. An Automatic Sleep Stage Classification Algorithm Using Improved Model Based Essence Features. Sensors 2020, 20, 4677. [Google Scholar] [CrossRef]
- Michielli, N.; Acharya, U.R.; Molinari, F. Cascaded LSTM recurrent neural network for automated sleep stage classification using single-channel EEG signals. Comput. Biol. Med. 2019, 106, 71–81. [Google Scholar] [CrossRef]
- Mousavi, S.; Afghah, F.; Acharya, R. SleepEEGNet: Automated sleep stage scoring with sequence to sequence deep learning approach. PLoS ONE 2019, 14, e0216456. [Google Scholar] [CrossRef]
- Sokolovsky, M.; Guerrero, F.; Paisarnsrisomsuk, S.; Ruiz, C.; Alvarez, S.A. Deep Learning for Automated Feature Discovery and Classification of Sleep Stages. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1835–1845. [Google Scholar] [CrossRef]
- Wei, Y.; Qi, X.; Wang, H.; Liu, Z.; Wang, G.; Yan, X. A Multi-Class Automatic Sleep Staging Method Based on Long Short-Term Memory Network Using Single-Lead Electrocardiogram Signals. IEEE Access 2019, 7, 85959–85970. [Google Scholar] [CrossRef]
- Urtnasan, E.; Park, J.-U.; Joo, E.Y.; Lee, K.-J. Deep Convolutional Recurrent Model for Automatic Scoring Sleep Stages Based on SingleLead ECG Signal. Diagnostics 2022, 12, 1235. [Google Scholar] [CrossRef] [PubMed]
- Toma, T.I.; Choi, S. An End-to-End Convolutional Recurrent Neural Network with Multi-Source Data Fusion for Sleep Stage Classification. In Proceedings of the 2023 International Conference on Artificial Intelligence in Information and Communication (ICAIIC), Bali, Indonesia, 20–23 February 2023; pp. 564–569. [Google Scholar]
- Durrant-Whyte, H.F. Sensor models and multisensor integration. In Autonomous Robot Vehicles; Springer: New York, NY, USA, 1990; pp. 73–89. [Google Scholar]
- Abdollahpour, M.; Rezaii, T.Y.; Farzamnia, A.; Saad, I. Transfer Learning Convolutional Neural Network for Sleep Stage Classification Using Two-Stage Data Fusion Framework. IEEE Access 2020, 8, 180618–180632. [Google Scholar] [CrossRef]
- Siami-Namini, S.; Tavakoli, N.; Namin, A.S. The Performance of LSTM and BiLSTM in Forecasting Time Series. In Proceedings of the 2019 IEEE International Conference on Big Data (Big Data), Los Angeles, CA, USA, 9–12 December 2019; pp. 3285–3292. [Google Scholar]
- Kemp, B.; Zwinderman, A.H.; Tuk, B.; Kamphuisen, H.A.; Oberye, J.J. Analysis of a sleep-dependent neuronal feedback loop: The slow-wave microcontinuity of the EEG. IEEE. Trans. Biomed. Eng. 2000, 47, 1185–1194. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.-K.; Stanley, H.E. Physiobank, physiotoolkit, and physionet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef]
- Sola, J.; Sevilla, J. Importance of input data normalization for the application of neural networks to complex industrial problems. IEEE Trans. Nuclear Sci. 1997, 44, 1464–1468. [Google Scholar] [CrossRef]
- Alam, M.M.; Rahman, M.H.; Ahmed, M.F.; Chowdhury, M.Z.; Jang, Y.M. Deep learning based optimal energy management for photovoltaic and battery energy storage integrated home micro-grid system. Sci. Rep. 2022, 12, 15133. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Jiang, A.; Liu, X.; Shang, J.; Zhang, L. LSTM-Based EEG Classification in Motor Imagery Tasks. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 2086–2095. [Google Scholar] [CrossRef]


| Sleep Stage | Symbol | Sleep EDF-20 | Sleep EDF-78 | ||
|---|---|---|---|---|---|
| Training Epochs | Test Epochs | Training Epochs | Test Epochs | ||
| Awake | W | 7042 | 1242 | 55,925 | 9869 |
| Stage 1 | S1 | 2384 | 420 | 18,248 | 3220 |
| Stage 2 | S2 | 15,129 | 2670 | 58,338 | 10,295 |
| Stage 3 and Stage 4 | S3 | 4848 | 855 | 11,042 | 1949 |
| Rapid Eye | REM | 6559 | 1158 | 21,901 | 3865 |
| Name of the Hyper- Parameters | Value |
|---|---|
| Training data shape | (3000, 1) |
| Iteration | 80 for pre-trained dual-channel modules and 40 for training the whole architecture |
| Optimizer | Adam |
| Batch size | 256 |
| Loss function | Categorical cross-entropy |
| Learning rate | 0.001 |
| PSG Channel | Overall Performance | Class-Wise Performance (F1 Score) (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Acc (%) | Kp | F1 score (%) | W | S1 | S2 | S3 | REM | |
| EEG Fpz-Cz + EOG and EEG Fpz-Cz + EMG | 91.44 | 0.89 | 88.09 | 94.83 | 69.83 | 93.23 | 94.26 | 88.30 |
| EEG Fpz-Cz + EMG and EEG Pz-Oz + EOG | 91.41 | 0.88 | 88.01 | 95.43 | 68.93 | 92.69 | 92.79 | 90.16 |
| EEG Fpz-Cz + EOG and EEG Pz-Oz + EMG | 91.01 | 0.88 | 87.39 | 95.01 | 67.92 | 93.26 | 93.66 | 87.13 |
| EEG Pz-Oz + EOG and EEG Pz-Oz + EMG | 88.49 | 0.84 | 84.43 | 92.74 | 62.66 | 90.59 | 89.09 | 87.09 |
| EEG Fpz-Cz + EOG and EEG Pz-Oz + EOG | 91.01 | 0.87 | 87.05 | 94.88 | 66.76 | 93.21 | 92.10 | 88.24 |
| EEG Fpz-Cz + EMG and EEG Pz-Oz + EMG | 89.75 | 0.85 | 85.95 | 93.51 | 64.92 | 91.62 | 91.02 | 88.67 |
| PSG Channel | Overall Performance | Class-Wise Performance | ||||||
|---|---|---|---|---|---|---|---|---|
| F1 Score (%) | ||||||||
| Acc (%) | Kp | F1 Score (%) | W | S1 | S2 | S3 | REM | |
| EEG Fpz-Cz + EOG and EEG Fpz-Cz + EMG | 89.94 | 0.86 | 86.65 | 95.48 | 68.76 | 91.49 | 90.02 | 87.50 |
| EEG Fpz-Cz + EMG and EEG Pz-Oz + EOG | 90.21 | 0.86 | 87.02 | 95.83 | 70.09 | 91.34 | 90.06 | 87.80 |
| EEG Fpz-Cz + EOG and EEG Pz-Oz + EMG | 89.51 | 0.85 | 86.18 | 95.73 | 68.98 | 90.92 | 89.66 | 85.59 |
| EEG Pz-Oz + EOG and EEG Pz-Oz + EMG | 83.56 | 0.77 | 0.78 | 93.57 | 51.34 | 85.94 | 81.74 | 75.22 |
| EEG Fpz-Cz + EOG and EEG Pz-Oz + EOG | 90.17 | 0.86 | 87.02 | 95.80 | 70.12 | 91.39 | 89.93 | 87.84 |
| EEG Fpz-Cz + EMG and EEG Pz-Oz + EMG | 89.69 | 0.85 | 86.46 | 95.69 | 69.68 | 91.18 | 90.09 | 85.67 |
| Paper | PSG Channel | Overall Performance | Class-Wise Performance (F1 Score) (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Acc (%) | Kp | F1 Score (%) | W | S1 | S2 | S3 | REM | ||
| Supratak et al. [11] | EEG Fpz-Cz | 82.0 | 76.9 | 76 | 84.7 | 46.6 | 85.9 | 84.8 | 82.4 |
| Phan et al. [12] | EEG Fpz- + EOG | 82.3 | 0.75 | 74.7 | - | - | - | - | - |
| Liu et al. [13] | EEG Fpz-Cz | 82.72 | 0.76 | 75.91 | 85 | 41 | 88 | 85 | 80 |
| EEG Pz-Oz | 80.99 | 0.73 | 72.69 | 85 | 33 | 87 | 82 | 78 | |
| Phan et al. [14] | EEG Fpz-Cz + EOG | 84.6 | 0.782 | 79.0 | 82.6 | 50.0 | 87.8 | 86.2 | 88.4 |
| Phan et al. [15] | EEG Fpz-Cz + EOG | 83.3 | 0.762 | 77.3 | - | - | - | - | - |
| Guillot et al. [16] | EEG Fpz-Cz + EEG Pz-Oz + EOG | - | - | 79.1 | - | - | - | - | - |
| Tianqi et al. [17] | EEG Fpz-Cz + EEG Pz-Oz + EOG + EMG | 85.8 | 0.80 | 81.2 | 92.3 | 54.5 | 87.8 | 85.4 | 85.9 |
| Xiaoqing et al. [20] | EEG Fpz-Cz + EEG Pz-Oz + EOG + EMG | 83.6 | 0.77 | 78.1 | 86.4 | 49.8 | 88.7 | 84.5 | 81.6 |
| Ours | EEG Fpz-Cz + EOG and EEG Fpz-Cz+EMG | 91.44 | 0.89 | 88.09 | 94.83 | 69.83 | 93.23 | 94.26 | 88.30 |
| Paper | PSG Channel | Overall Performance | Class-Wise Performance (F1 Score) (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Acc (%) | Kp (%) | F1 Score (%) | W | S1 | S2 | S3 | REM | ||
| Phan et al. [15] | EEG Fpz-Cz + EOG | 80.6 | 0.728 | 76.7 | - | - | - | - | - |
| Guillot et al. [16] | EEG Fpz-Cz + EEG Pz-Oz + EOG | - | - | 76.3 | - | - | - | - | - |
| Khalili et al. [18] | EEG Fpz-Cz | 82.46 | 0.76 | 76.14 | 92.4 | 48.1 | 84.6 | 73.8 | 81.6 |
| EEG Pz-Oz | 79.33 | 0.71 | 76.4 | 87.1 | 53.0 | 79.8 | 72.5 | 74.2 | |
| Eldele et al. [19] | EEG Fpz-Cz | 81.3 | 0.74 | 75.1 | 92.0 | 42.0 | 85.0 | 82.1 | 74.2 |
| Mousavi et al. [23] | EEG Fpz-Cz | 80.03 | 0.73 | 73.55 | 91.72 | 44.05 | 82.49 | 73.45 | 76.06 |
| EEG Pz-Oz | 77.56 | 68.94 | 70.00 | 90.26 | 42.21 | 79.71 | 94.83 | 72.19 | |
| Ours | EEG Fpz-Cz + EMG and EEG Pz-Oz + EOG | 90.21 | 0.86 | 87.02 | 95.83 | 70.09 | 91.34 | 90.06 | 87.80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toma, T.I.; Choi, S. An End-to-End Multi-Channel Convolutional Bi-LSTM Network for Automatic Sleep Stage Detection. Sensors 2023, 23, 4950. https://doi.org/10.3390/s23104950
Toma TI, Choi S. An End-to-End Multi-Channel Convolutional Bi-LSTM Network for Automatic Sleep Stage Detection. Sensors. 2023; 23(10):4950. https://doi.org/10.3390/s23104950
Chicago/Turabian StyleToma, Tabassum Islam, and Sunwoong Choi. 2023. "An End-to-End Multi-Channel Convolutional Bi-LSTM Network for Automatic Sleep Stage Detection" Sensors 23, no. 10: 4950. https://doi.org/10.3390/s23104950
APA StyleToma, T. I., & Choi, S. (2023). An End-to-End Multi-Channel Convolutional Bi-LSTM Network for Automatic Sleep Stage Detection. Sensors, 23(10), 4950. https://doi.org/10.3390/s23104950






