Transformer-Based Approach to Melanoma Detection
Abstract
:1. Introduction
- The proposed model is trained and tested on public skin cancer data from the ISIC challenge, making a fair comparison with competitors with similar approaches to dealing with the same clinical issue possible;
- The performance of the presented ViT-based architecture for skin lesion image classification exceeds that of the state of the art by modeling long-range spatial relationships;
- Extensive tuning studies are performed to investigate the effect of individual elements of the ViT model.
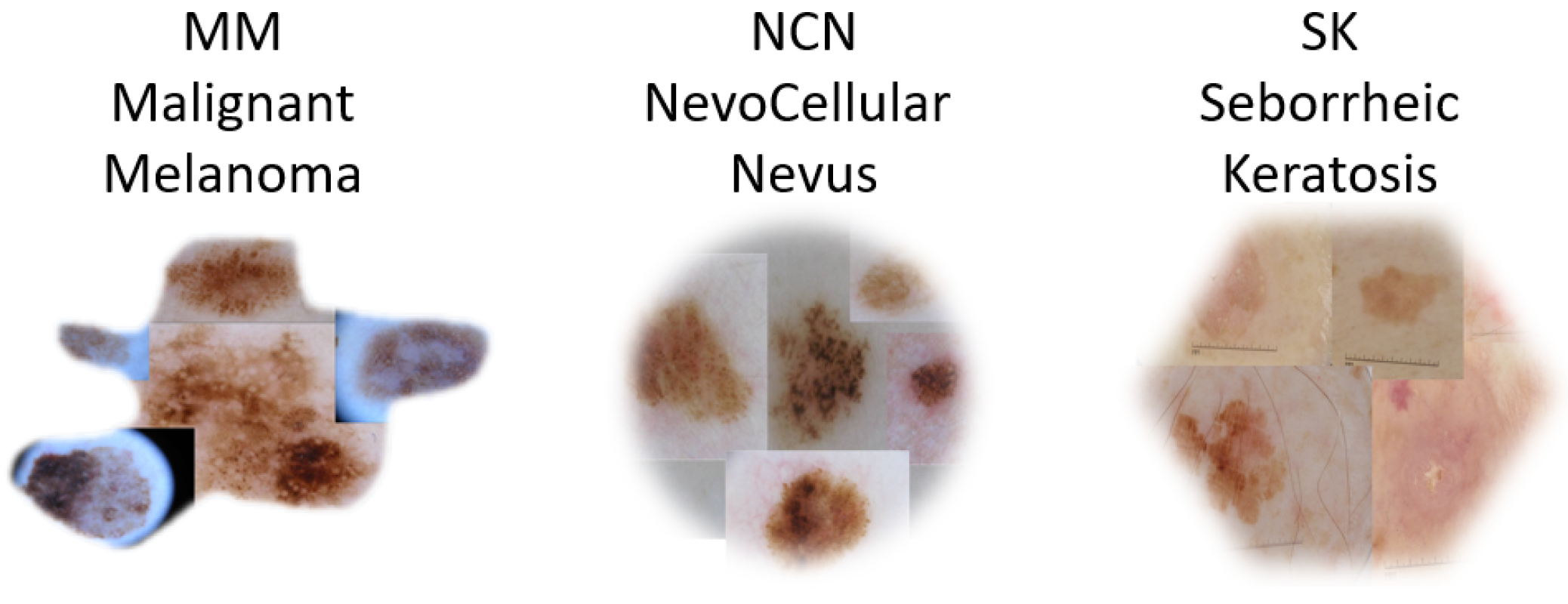
2. Dataset
3. Methodology
3.1. The End-to-End Workflow
- Input image: The input image was fed to the model at its original size. A blue-colored block represented the workflow’s starting point.
- Resizing: The images were resized to 384 × 384 in order to be properly processed by the model.
- Random resized cropping: The training images were randomly resized and cropped as a part of the data augmentation procedure. This block was marked with yellow to indicate that it only operated on the training images.
- RandAugment: The RandAugment library [23] was applied to the training set for data augmentation purposes. This block, as with the previous one, was marked in yellow, with the exact same meaning.
- Patching: The original image was divided into equally sized, non-overlapping 32 × 32 square patches.
- Flattening: Each patch was vectorized (flattening) in order to have an embedding for the corresponding token in the input sequence.
- Embedding + positional encoding: The patches were encoded to account for their positional information.
- Concatenation (input sequence): The positional encoded vector was added to the corresponding token.
- Layer normalization: The resulting vector was layer normalized and fed to the transformer encoder.
- Multi-head attention: The multi-head attention mechanism came into play, reproducing the behavior of multiple “experts” analyzing different areas of the picture, exchanging information, and interacting to come up with a coherent interpretation of the image.
- Layer normalization: The output of the multi-head attention blocks was added to the embedded input by means of a skip connection, and it was later normalized once more.
- Multilayer perceptron: The resulting output was fed to an MLP consisting of two fully connected layers with a GELU activation function [24].
- Classification head: The MLP output was added to the multi-head attention output through another skip connection. The classification head is a fully connected layer that will produce the model output.
- Classification output: The final outcome was a value that would be used to determine which class the input image belonged to. This block, highlighted in green, marks the end of the workflow.
Multi-Head Attention
3.2. Experimental Set-Up
Evaluation Metrics
4. Experimental Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| AUROC | Area under the receiver operating characteristic |
| CAD | Computer-aided diagnosis |
| CNNs | Convolutional neural networks |
| DL | Deep learning |
| e-Health | Electronic Health |
| FTNs | Fully Transformer networks |
| GANs | Generative adversarial networks |
| GELU | Gaussian error linear unit |
| IRNNs | Identity recurrent neural networks |
| ISIC | International Skin Imaging Collaboration |
| LSTM | Long short-term memory |
| m-Health | Mobile Health |
| ML | Machine learning |
| MM | Malignant melanoma |
| MLP | Multilayer perceptron |
| NCN | Nevocellular nevus |
| RCNNs | Recurrent convolutional neural networks |
| RNNs | Recurrent neural networks |
| SK | Seborrheic keratosis |
| SPTs | Spatial pyramid transformers |
| UV | Ultraviolet |
| ViTs | Vision transformers |
| WHO | World Health Organization |
References
- Hu, W.; Fang, L.; Ni, R.; Zhang, H.; Pan, G. Changing trends in the disease burden of non-melanoma skin cancer globally from 1990 to 2019 and its predicted level in 25 years. BMC Cancer 2022, 22, 836. [Google Scholar] [CrossRef] [PubMed]
- Lacey, J.V., Jr.; Devesa, S.S.; Brinton, L.A. Recent trends in breast cancer incidence and mortality. Environ. Mol. Mutagen. 2002, 39, 82–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uong, A.; Zon, L.I. Melanocytes in development and cancer. J. Cell. Physiol. 2010, 222, 38–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [Green Version]
- Verma, R.; Anand, S.; Vaja, C.; Bade, R.; Shah, A.; Gaikwad, K. Metastatic malignant melanoma: A case study. Int. J. Sci. Study 2016, 4, 188–190. [Google Scholar]
- Naik, P.P. Cutaneous malignant melanoma: A review of early diagnosis and management. World J. Oncol. 2021, 12, 7. [Google Scholar] [CrossRef]
- Jutzi, T.B.; Krieghoff-Henning, E.I.; Holland-Letz, T.; Utikal, J.S.; Hauschild, A.; Schadendorf, D.; Sondermann, W.; Fröhling, S.; Hekler, A.; Schmitt, M.; et al. Artificial intelligence in skin cancer diagnostics: The patients’ perspective. Front. Med. 2020, 7, 233. [Google Scholar] [CrossRef]
- Pollastri, F.; Parreño, M.; Maroñas, J.; Bolelli, F.; Paredes, R.; Ramos, D.; Grana, C. A deep analysis on high-resolution dermoscopic image classification. IET Comput. Vis. 2021, 15, 514–526. [Google Scholar] [CrossRef]
- Lucieri, A.; Bajwa, M.N.; Braun, S.A.; Malik, M.I.; Dengel, A.; Ahmed, S. ExAID: A multimodal explanation framework for computer-aided diagnosis of skin lesions. Comput. Methods Programs Biomed. 2022, 215, 106620. [Google Scholar] [CrossRef]
- Pollastri, F.; Bolelli, F.; Paredes, R.; Grana, C. Augmenting data with GANs to segment melanoma skin lesions. Multimed. Tools Appl. 2020, 79, 15575–15592. [Google Scholar] [CrossRef] [Green Version]
- Aljohani, K.; Turki, T. Automatic Classification of Melanoma Skin Cancer with Deep Convolutional Neural Networks. Ai 2022, 3, 512–525. [Google Scholar] [CrossRef]
- Allugunti, V.R. A machine learning model for skin disease classification using convolution neural network. Int. J. Comput. Program. Database Manag. 2022, 3, 141–147. [Google Scholar] [CrossRef]
- Malo, D.C.; Rahman, M.M.; Mahbub, J.; Khan, M.M. Skin Cancer Detection using Convolutional Neural Network. In Proceedings of the 2022 IEEE 12th Annual Computing and Communication Workshop and Conference (CCWC), Virtual Conference, 26–29 January 2022; pp. 169–176. [Google Scholar]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, M.; Hu, X. Recurrent convolutional neural network for object recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3367–3375. [Google Scholar]
- Chandra, B.; Sharma, R.K. On improving recurrent neural network for image classification. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AL, USA, 14–19 May 2017; pp. 1904–1907. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. [Google Scholar]
- Bhojanapalli, S.; Chakrabarti, A.; Glasner, D.; Li, D.; Unterthiner, T.; Veit, A. Understanding robustness of transformers for image classification. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10231–10241. [Google Scholar]
- Lanchantin, J.; Wang, T.; Ordonez, V.; Qi, Y. General multi-label image classification with transformers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 16478–16488. [Google Scholar]
- Xie, J.; Wu, Z.; Zhu, R.; Zhu, H. Melanoma detection based on swin transformer and SimAM. In Proceedings of the 2021 IEEE 5th Information Technology, Networking, Electronic and Automation Control Conference (ITNEC), Xi’an, China, 15–17 October 2021; Volume 5, pp. 1517–1521. [Google Scholar]
- Roy, V.K.; Thakur, V.; Goyal, N. Vision Transformer Framework Approach for Melanoma Skin Disease Identification. 2023. Available online: https://assets.researchsquare.com/files/rs-2536632/v1/00ee7438-9206-4cfd-a8ad-319813d22bb8.pdf?c=1682720069 (accessed on 17 May 2023).
- Codella, N.C.F.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2018, arXiv:1710.05006. [Google Scholar]
- Cubuk, E.D.; Zoph, B.; Shlens, J.; Le, Q.V. RandAugment: Practical automated data augmentation with a reduced search space. arXiv 2019, arXiv:1909.13719. [Google Scholar]
- Hendrycks, D.; Gimpel, K. Gaussian Error Linear Units (GELUs). arXiv 2020, arXiv:1606.08415. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2019, arXiv:1810.04805. [Google Scholar]
- Florkowski, C.M. Sensitivity, specificity, receiver-operating characteristic (ROC) curves and likelihood ratios: Communicating the performance of diagnostic tests. Clin. Biochem. Rev. 2008, 29, S83. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef] [Green Version]
- Gulzar, Y.; Khan, S.A. Skin Lesion Segmentation Based on Vision Transformers and Convolutional Neural Networks—A Comparative Study. Appl. Sci. 2022, 12, 5990. [Google Scholar] [CrossRef]
- He, X.; Tan, E.L.; Bi, H.; Zhang, X.; Zhao, S.; Lei, B. Fully transformer network for skin lesion analysis. Med. Image Anal. 2022, 77, 102357. [Google Scholar] [CrossRef] [PubMed]
- Azad, R.; Asadi-Aghbolaghi, M.; Fathy, M.; Escalera, S. Attention deeplabv3+: Multi-level context attention mechanism for skin lesion segmentation. In Proceedings of the Computer Vision—ECCV 2020 Workshops, Glasgow, UK, 23–28 August 2020; Proceedings, Part I 16. Springer: Berlin/Heidelberg, Germany, 2020; pp. 251–266. [Google Scholar]
- Wu, W.; Mehta, S.; Nofallah, S.; Knezevich, S.; May, C.J.; Chang, O.H.; Elmore, J.G.; Shapiro, L.G. Scale-aware transformers for diagnosing melanocytic lesions. IEEE Access 2021, 9, 163526–163541. [Google Scholar] [CrossRef] [PubMed]
- Aladhadh, S.; Alsanea, M.; Aloraini, M.; Khan, T.; Habib, S.; Islam, M. An effective skin cancer classification mechanism via medical vision transformer. Sensors 2022, 22, 4008. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.K.; Shaikh, M.A.; Srihari, S.N.; Gao, M. Soft-Attention Improves Skin Cancer Classification Performance. medRxiv 2021. [Google Scholar] [CrossRef]
- Zhang, B.; Jin, S.; Xia, Y.; Huang, Y.; Xiong, Z. Attention Mechanism Enhanced Kernel Prediction Networks for Denoising of Burst Images. arXiv 2019, arXiv:1910.08313. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Wang, F.; Jiang, M.; Qian, C.; Yang, S.; Li, C.; Zhang, H.; Wang, X.; Tang, X. Residual Attention Network for Image Classification. arXiv 2017, arXiv:1704.06904. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]





| Hyperparameter | Value |
|---|---|
| Number of layers | 24 |
| Number of heads | 16 |
| Hidden size | 1024 |
| Optimizer | Adam with = 0.99, = 0.999, |
| Learning rate | |
| Learning rate scheduler | linear with a warm-up ratio of 0.1 |
| Batch size | 16 |
| Gradient accumulation steps | 4 |
| Configuration | Batch Size | Learning Rate | Layers | Accuracy |
|---|---|---|---|---|
| #1 | 16 | 24 | 0.948 | |
| #2 (Best Model) | 16 | 24 | 0.948 | |
| #3 | 16 | 24 | 0.948 | |
| #4 | 16 | 24 | 0.945 | |
| #5 | 16 | 20 | 0.925 | |
| #6 | 16 | 16 | 0.902 | |
| #7 | 16 | 12 | 0.850 | |
| #8 | 16 | 10 | 0.817 | |
| #9 | 16 | 8 | 0.788 | |
| #10 | 16 | 4 | 0.707 | |
| #11 | 16 | 2 | 0.655 |
| Model Name | Accuracy | Sensitivity | Specificity | AUROC |
|---|---|---|---|---|
| IRv2 + soft attention [33] | 0.904 | 0.916 | 0.833 | 0.959 |
| ARL-CNN50 [34] | 0.868 | 0.878 | 0.867 | 0.958 |
| SEnet50 [35] | 0.863 | 0.856 | 0.865 | 0.952 |
| RAN50 [36] | 0.862 | 0.878 | 0.859 | 0.942 |
| ResNet50 [37] | 0.842 | 0.867 | 0.837 | 0.948 |
| Proposed ViT-based approach | 0.948 | 0.928 | 0.967 | 0.948 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cirrincione, G.; Cannata, S.; Cicceri, G.; Prinzi, F.; Currieri, T.; Lovino, M.; Militello, C.; Pasero, E.; Vitabile, S. Transformer-Based Approach to Melanoma Detection. Sensors 2023, 23, 5677. https://doi.org/10.3390/s23125677
Cirrincione G, Cannata S, Cicceri G, Prinzi F, Currieri T, Lovino M, Militello C, Pasero E, Vitabile S. Transformer-Based Approach to Melanoma Detection. Sensors. 2023; 23(12):5677. https://doi.org/10.3390/s23125677
Chicago/Turabian StyleCirrincione, Giansalvo, Sergio Cannata, Giovanni Cicceri, Francesco Prinzi, Tiziana Currieri, Marta Lovino, Carmelo Militello, Eros Pasero, and Salvatore Vitabile. 2023. "Transformer-Based Approach to Melanoma Detection" Sensors 23, no. 12: 5677. https://doi.org/10.3390/s23125677
APA StyleCirrincione, G., Cannata, S., Cicceri, G., Prinzi, F., Currieri, T., Lovino, M., Militello, C., Pasero, E., & Vitabile, S. (2023). Transformer-Based Approach to Melanoma Detection. Sensors, 23(12), 5677. https://doi.org/10.3390/s23125677










