An Optical Sensory System for Assessment of Residual Cancer Burden in Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy
Abstract
:1. Introduction
2. Materials and Methods
2.1. NIR Opti-Scan Probe for Breast Tissue Imaging and Cancer Detection
2.2. Study Desing and Scanning Procedure
2.3. Machine Learning Models for Breast Optical Properties and Residual Cancer Burden
Review of Current ML Methods for Breast Optical Properties and Residual Cancer Burden
2.4. Proposed ML Model for Breast Optical Properties and Residual Cancer Burden
3. Results: Machine-Learning-Based Method for Breast Tissue Optical Property Determination and Treatment Response Monitoring
3.1. Optical Property Determination
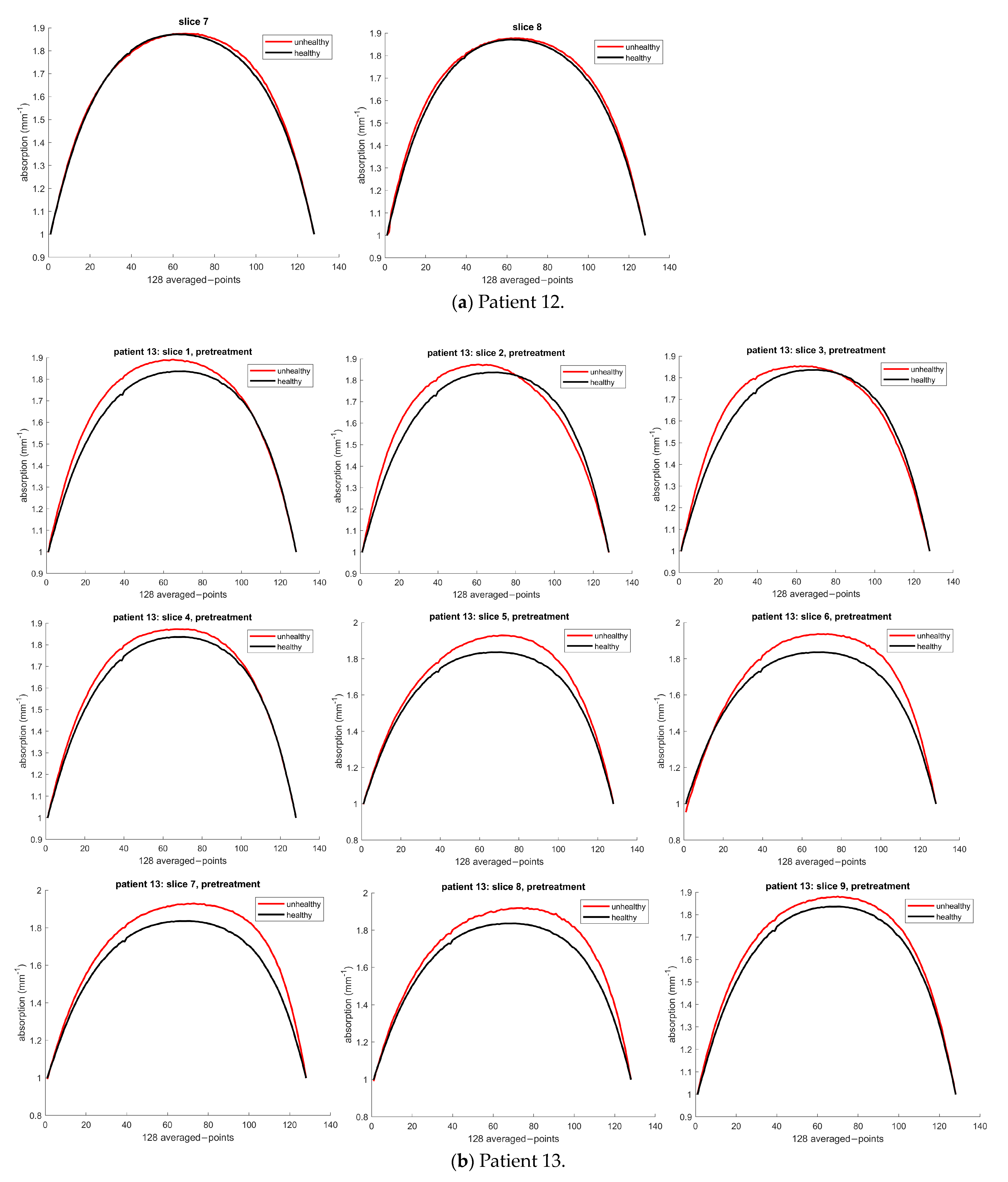
3.1.1. Patient 12
3.1.2. Patient 13
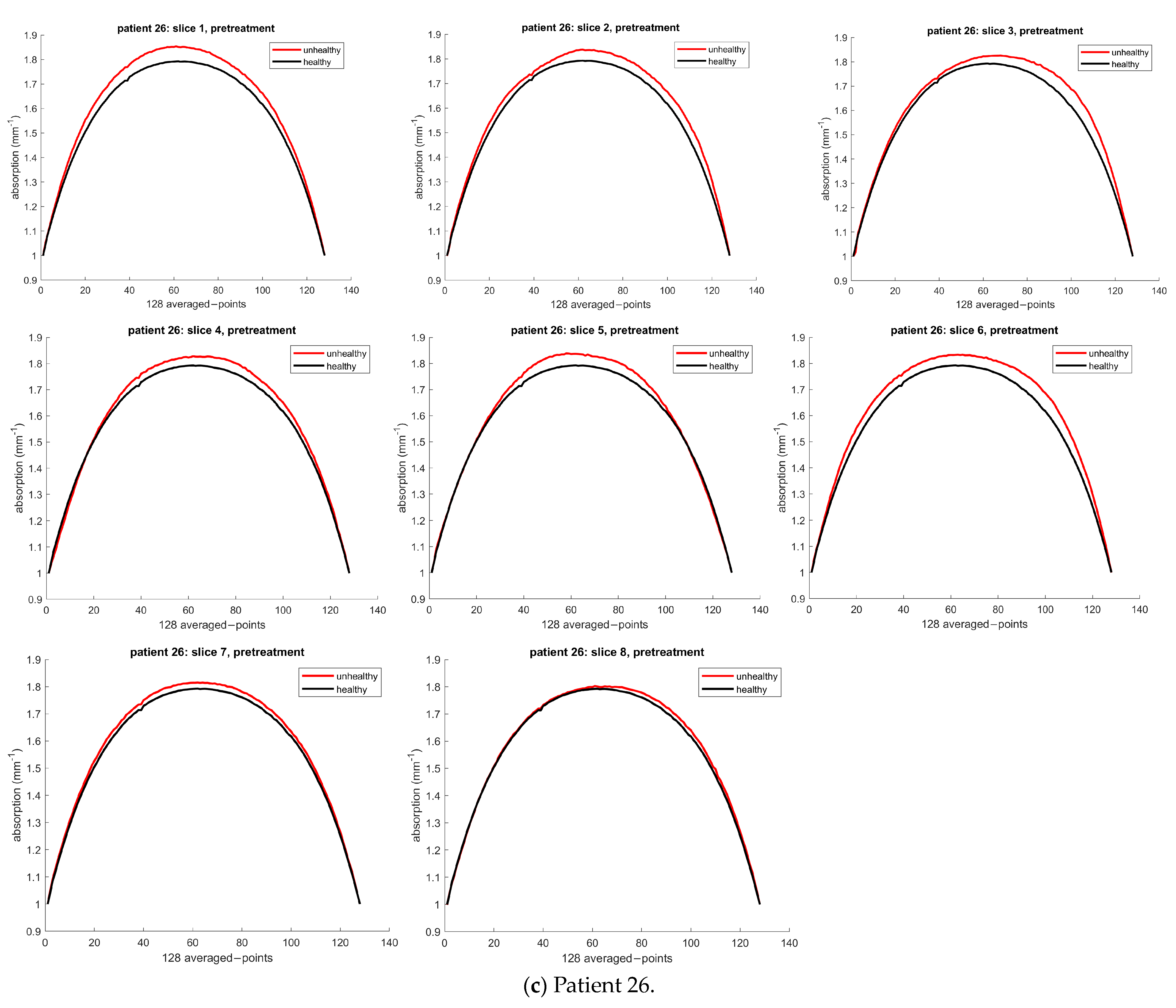
3.1.3. Patient 26
3.2. Treatment Response Monitoring and Residual Cancer Burden (RCB)
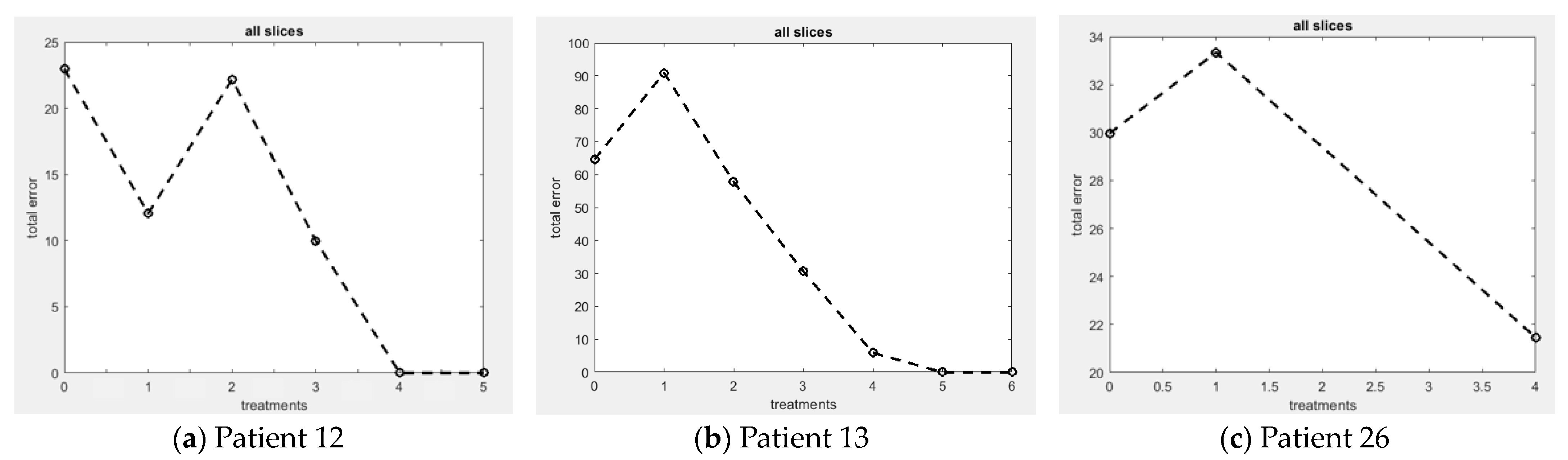
3.2.1. Patient 12
3.2.2. Patient 13
3.2.3. Patient 26
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, M.; von Minckwitz, G.; Mamounas, E.P.; Cameron, D.; Carey, L.A.; Cristofanilli, M.; Denkert, C.; Eiermann, W.; Gnant, M.; Harris, J.R.; et al. Recommendations from an International Consensus Conference on the Current Status and Future of Neoadjuvant Systemic Therapy in Primary Breast Cancer. Ann. Surg. Oncol. 2012, 19, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- Cortadellas, T.; Argacha, P.; Acosta, J.; Rabasa, J.; Peiró, R.; Gomez, M.; Rodellar, L.; Gomez, S.; Navarro-Golobart, A.; Sanchez-Mendez, S.; et al. Estimation of tumor size in breast cancer comparing clinical examination, mammography, ultrasound and MRI—Correlation with the pathological analysis of the surgical specimen. Gland. Surg. 2017, 6, 330–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef] [Green Version]
- van der Hage, J.H.; van de Velde, C.C.; Mieog, S.J.; Charehbili, A. Preoperative chemotherapy for women with operable breast cancer. Cochrane Database Syst. Rev. 2007, 2007, CD005002. [Google Scholar] [CrossRef]
- Masuda, N.; Lee, S.-J.; Ohtani, S.; Im, Y.-H.; Lee, E.-S.; Yokota, I.; Kuroi, K.; Im, S.-A.; Park, B.-W.; Kim, S.-B.; et al. Adjuvant Capecitabine for Breast Cancer after Preoperative Chemotherapy. N. Engl. J. Med. 2017, 376, 2147–2159. [Google Scholar] [CrossRef]
- Yau, C.; Osdoit, M.; van der Noordaa, M.; Shad, S.; Wei, J.; de Croze, D.; Hamy, A.-S.; Laé, M.; Reyal, F.; Sonke, G.S.; et al. Residual cancer burden after neoadjuvant chemotherapy and long-term survival outcomes in breast cancer: A multicentre pooled analysis of 5161 patients. Lancet Oncol. 2022, 23, 149–160. [Google Scholar] [CrossRef]
- Pinard, C.; Debled, M.; Ben Rejeb, H.; Velasco, V.; de Lara, C.T.; Hoppe, S.; Richard, E.; Brouste, V.; Bonnefoi, H.; MacGrogan, G. Residual cancer burden index and tumor-infiltrating lymphocyte subtypes in triple-negative breast cancer after neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2020, 179, 11–23. [Google Scholar] [CrossRef]
- Symmans, W.F.; Peintinger, F.; Hatzis, C.; Rajan, R.; Kuerer, H.; Valero, V.; Assad, L.; Poniecka, A.; Hennessy, B.; Green, M.; et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J. Clin. Oncol. 2007, 25, 4414–4422. [Google Scholar] [CrossRef]
- Shokoufi, M.; Haeri, Z.; Lim, Z.Y.; Ramaseshan, R.; Golnaraghi, F. Translation of a portable diffuse optical breast scanner probe for clinical application: A preliminary study. Biomed. Phys. Eng. Express 2020, 6, 015037. [Google Scholar] [CrossRef]
- Cong, W.; Intes, X.; Wang, G. Optical tomographic imaging for breast cancer detection. J. Biomed. Opt. 2017, 22, 096011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tromberg, B.J.; Shah, N.; Lanning, R.; Cerussi, A.; Espinoza, J.; Pham, T.; Svaasand, L.; Butler, J. Non-invasive in vivo characterization of breast tumors using photon migration spectroscopy. Neoplasia 2000, 2, 26–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ntziachristos, V.; Yodh, A.G.; Schnall, M.; Chance, B. Concurrent MRI and diffuse optical tomography of breast after indocyanine green enhancement. Proc. Natl. Acad. Sci. USA 2000, 97, 2767–2772. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.; Kurtzman, S.H.; Hegde, P.; Tannenbaum, S.; Kane, M.; Huang, M.; Chen, N.G.; Jagjivan, B.; Zarfos, K. Utilizing optical tomography with ultrasound localization to image heterogeneous hemoglobin distribution in large breast cancers. Neoplasia 2005, 7, 263–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vavadi, H.; Mostafa, A.; Zhou, F.; Uddin, K.M.S.; Althobaiti, M.; Xu, C.; Bansal, R.; Ademuyiwa, F.; Poplack, S.; Zhu, Q. Compact ultrasound-guided diffuse optical tomography system for breast cancer imaging. J. Biomed. Opt. 2018, 24, 021203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, G. Brief review on learning-based methods for optical tomography. J. Innov. Opt. Health Sci. 2019, 12, 1930011. [Google Scholar] [CrossRef] [Green Version]
- Momtahen, S.; Shokoufi, M.; Ramaseshan, R.; Golnaraghi, F. Near-infrared Probe and Imaging System for Breast Cancer Tumor Localization Application. IEEE Can. J. Electr. Comput. Eng. 2023. [Google Scholar]
- Momtahen, S.; Ramaseshan, R.; Golnaraghi, F. Nonlinear Regression-Based Functional Image Reconstruction for Breast Cancer Diagnosis and Treatment Monitoring using Diffuse Optical Probe. Biomed. Phys. Eng. Express 2023. submitted. [Google Scholar]
- Momtahen, S.; Momtahen, M.; Ramaseshan, R.; Golnaraghi, F. A Machine Learning Approach: NIR Scattering Data Analysis for Breast Cancer Detection and Classification. In Proceedings of the 1st IEEE Industrial Electronics Society Annual Conference, Kharagpur, India, 9–11 December 2022; p. 1085. Available online: https://ies-oncon.com/OnConPapers2022.pdf (accessed on 18 June 2023).
- Momtahen, M.; Momtahen, S.; Remaseshan, R.; Golnaraghi, F.; Engineering, S. Early Detection of Breast Cancer using Diffuse Optical Probe and Ensemble Learning Method. In Proceedings of the IEEE MTT-S International Conference on Numerical Electromagnetic and Multiphysics Modeling and Optimization, Winnipeg, Canada, 28–30 June 2023. [Google Scholar]
- Shokoufi, M.; Golnaraghi, F. Handheld diffuse optical breast scanner probe for cross-sectional imaging of breast tissue. J. Innov. Opt. Health Sci. 2019, 12, 1950008. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Raghuram, A.; Kim, H.K.; Hielscher, A.H.; Robinson, J.T.; Veeraraghavan, A. High Resolution, Deep Imaging Using Confocal Time-of-Flight Diffuse Optical Tomography. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 2206–2219. Available online: http://arxiv.org/abs/2101.11680 (accessed on 18 June 2023). [CrossRef]
- Ohmae, E.; Yoshizawa, N.; Yoshimoto, K.; Hayashi, M.; Wada, H.; Mimura, T.; Asano, Y.; Ogura, H.; Yamashita, Y.; Sakahara, H.; et al. Comparison of lipid and water contents by time-domain diffuse optical spectroscopy and dual-energy computed tomography in breast cancer patients. Appl. Sci. 2019, 9, 1482. [Google Scholar] [CrossRef] [Green Version]
- Ben Yedder, H.; Cardoen, B.; Shokoufi, M.; Golnaraghi, F.; Hamarneh, G. Multitask deep learning reconstruction and localization of lesions in limited angle diffuse optical tomography. IEEE Trans. Med. Imaging 2022, 41, 515–530. [Google Scholar] [CrossRef]
- Hoshi, Y.; Yamada, Y. Overview of diffuse optical tomography and its clinical applications. J. Biomed. Opt. 2016, 21, 091312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belagiannis, V.; Rupprecht, C.; Carneiro, G.; Navab, N. Robust optimization for deep regression. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; Volume 2015, pp. 2830–2838. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Sun, Q.; Li, Z.; Sun, Z.; Jia, K. Back-propagation neural network-based reconstruction algorithm for diffuse optical tomography. J. Biomed. Opt. 2018, 24, 051407. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.; Sabir, S.; Heo, D.; Kim, K.H.; Wahab, A.; Choi, Y.; Lee, S.-I.; Chae, E.Y.; Kim, H.H.; Bae, Y.M.; et al. Deep Learning Diffuse Optical Tomography. IEEE Trans. Med. Imaging 2020, 39, 877–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahmassebi, A.; Wengert, G.J.; Helbich, T.H.; Bago-Horvath, Z.; Alaei, S.; Bartsch, R.; Dubsky, P.; Baltzer, P.; Clauser, P.; Kapetas, P.; et al. Impact of Machine Learning With Multiparametric Magnetic Resonance Imaging of the Breast for Early Prediction of Response to Neoadjuvant Chemotherapy and Survival Outcomes in Breast Cancer Patients. Invest. Radiol. 2019, 54, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Janse, M.H.A.; Janssen, L.M.; van der Velden, B.H.M.; Moman, M.R.; der Ben, E.J.M.W.; Kock, M.C.J.M.; Viergever, M.A.; van Diest, P.J.; Gilhuijs, K.G.A. Deep Learning-Based Segmentation of Locally Advanced Breast Cancer on MRI in Relation to Residual Cancer Burden: A Multiinstitutional Cohort Study. J. Magn. Reson. Imaging 2023. [Google Scholar] [CrossRef]
- Janssen, B.V.; Theijse, R.; van Roessel, S.; de Ruiter, R.; Berkel, A.; Huiskens, J.; Busch, O.R.; Wilmink, J.W.; Kazemier, G.; Valkema, P.; et al. Artificial Intelligence-Based Segmentation of Residual Tumor in Histopathology of Pancreatic Cancer after Neoadjuvant Treatment. Cancers 2021, 13, 5089. [Google Scholar] [CrossRef]
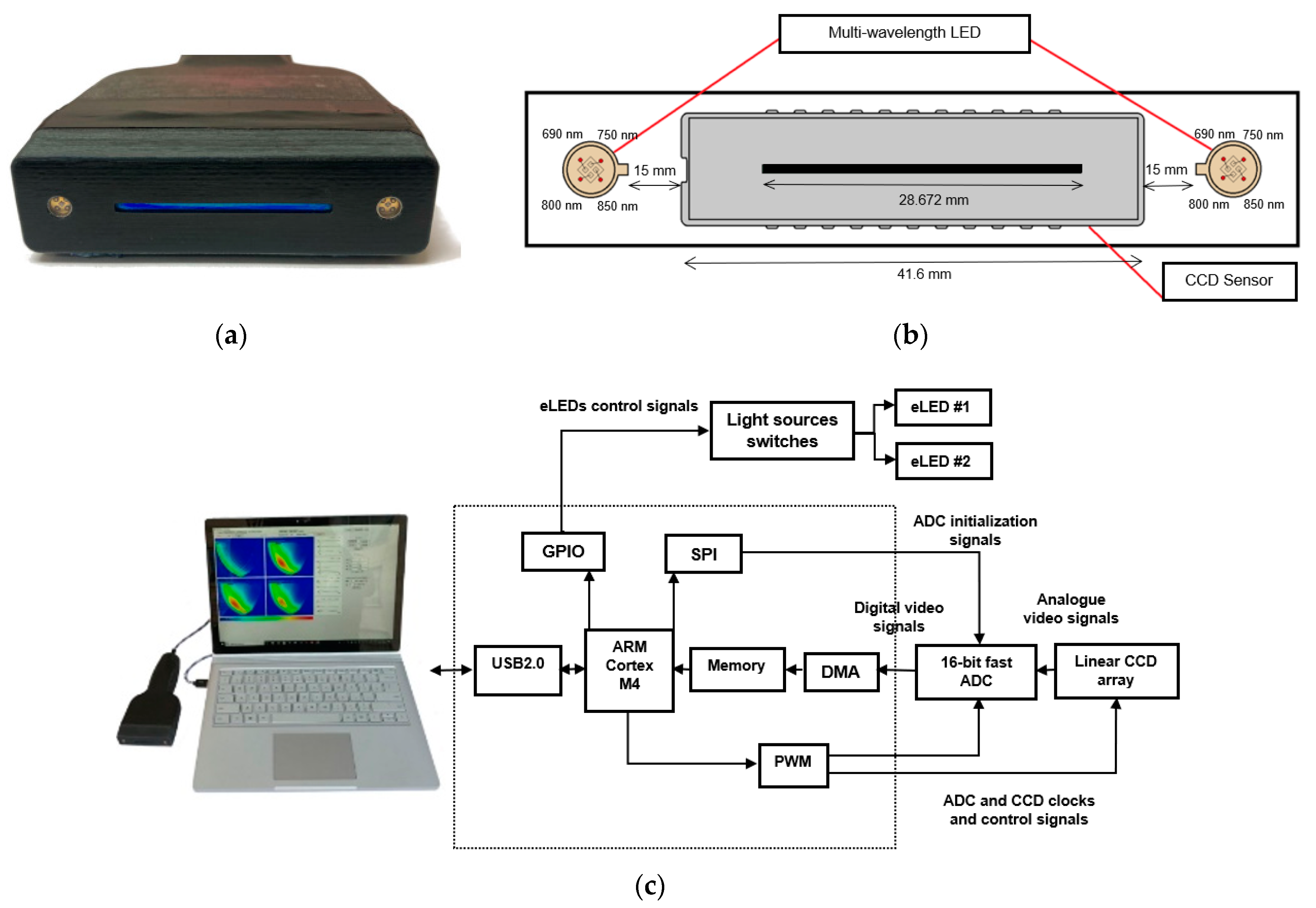
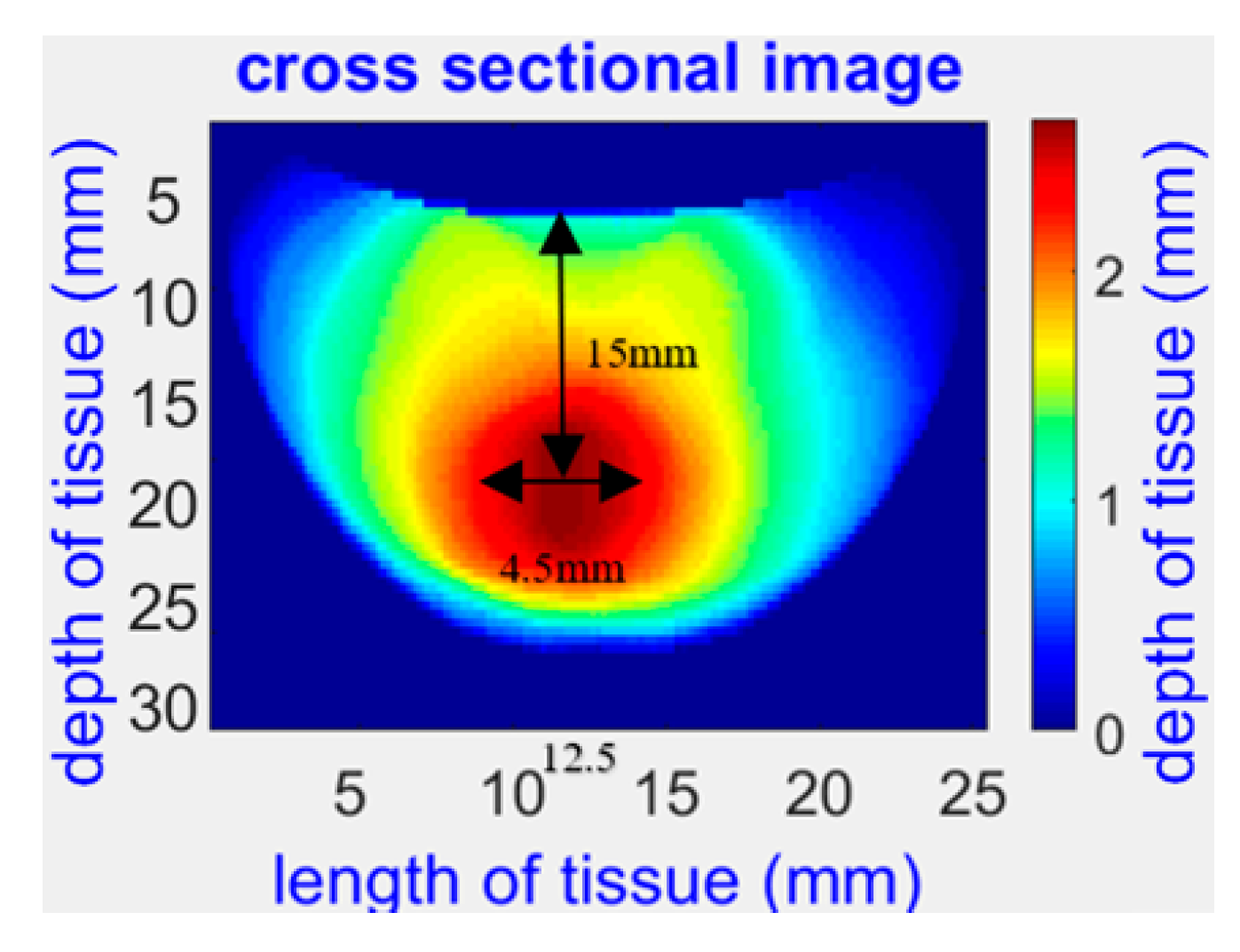
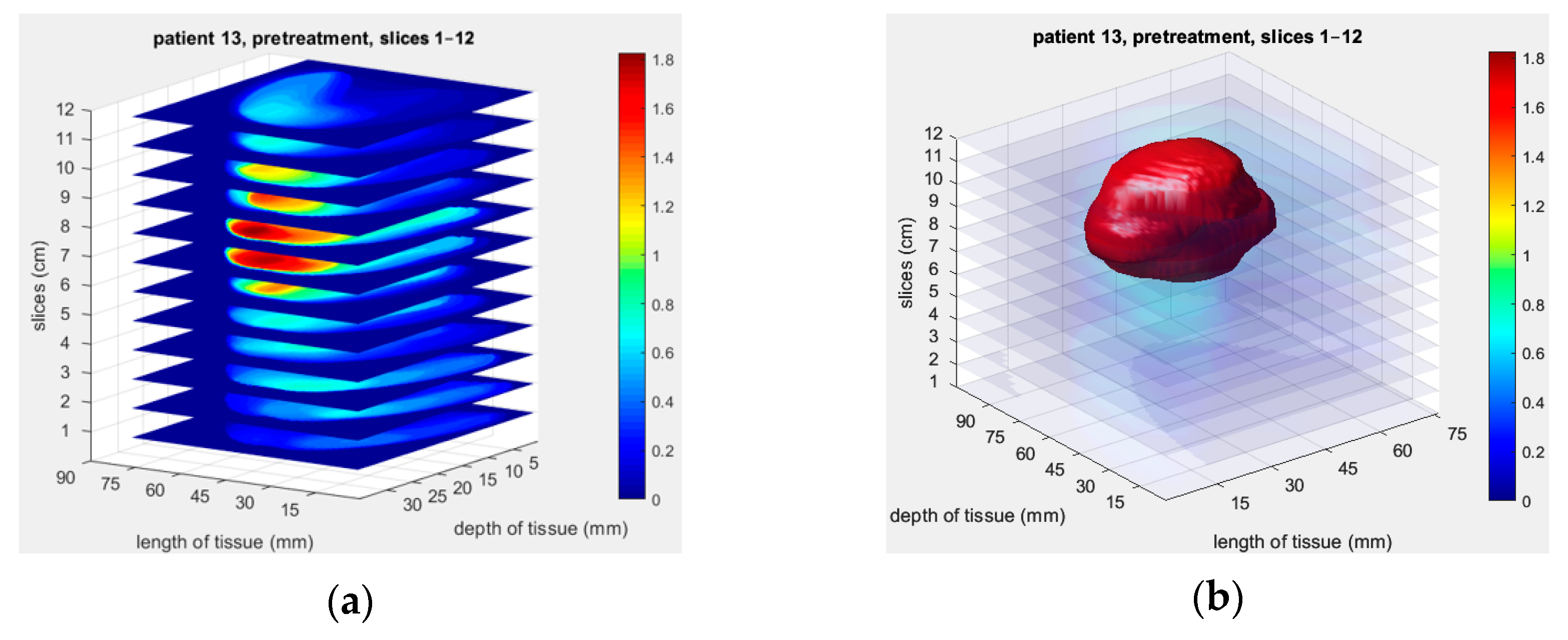
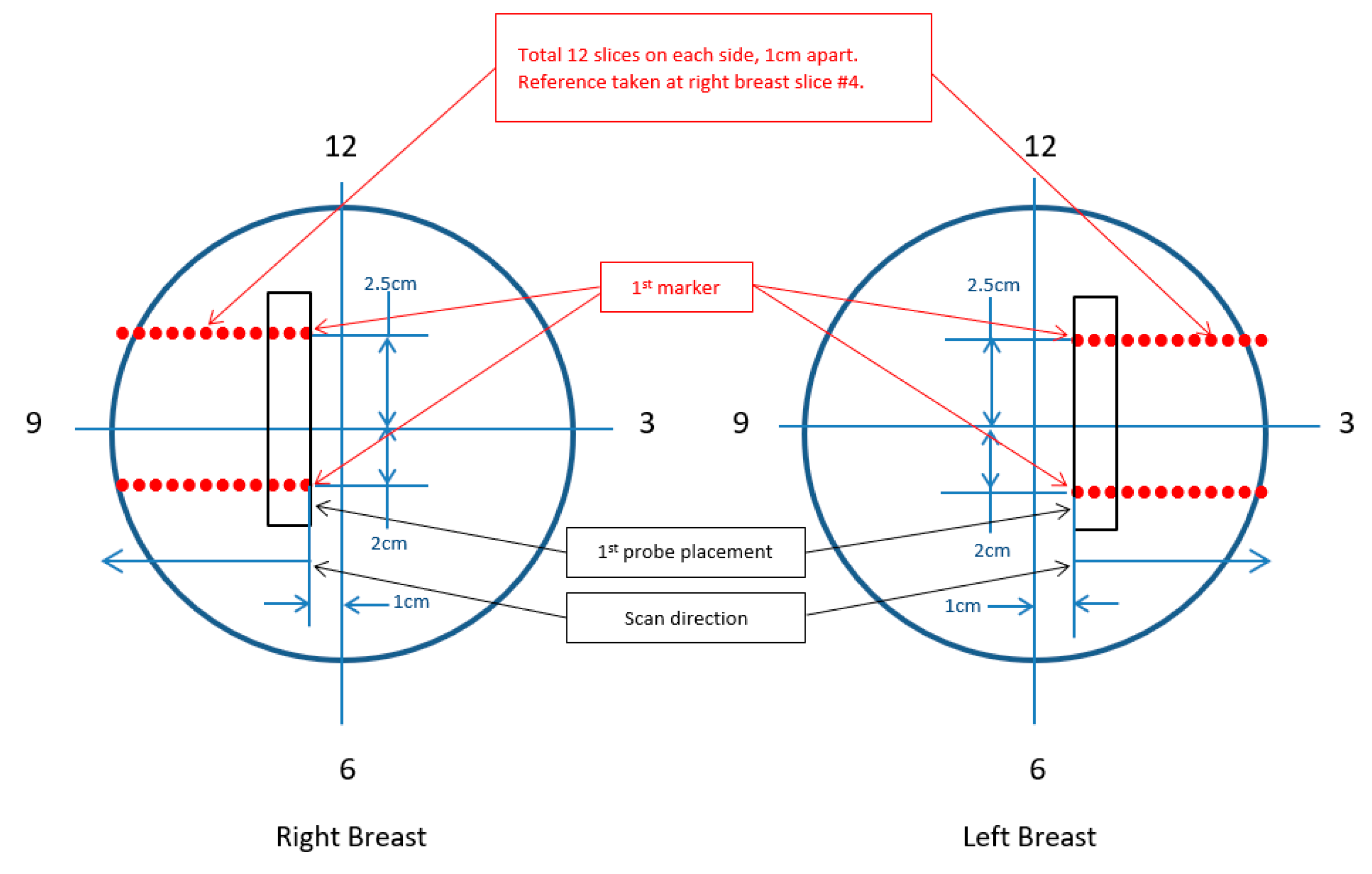
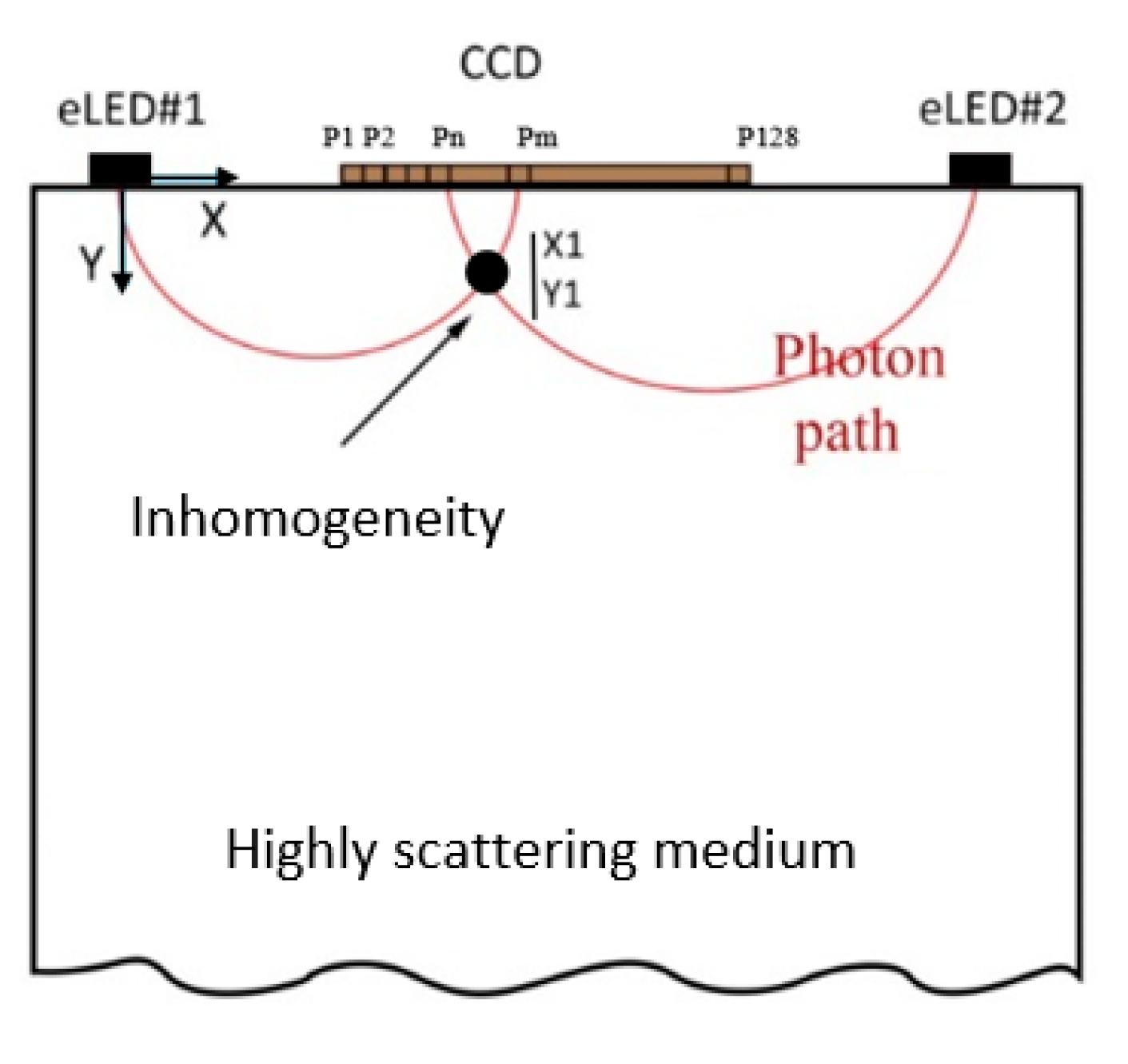


| Parameter | Value |
|---|---|
| Imaging technology | Near-infrared optical imaging |
| Detector type | Linear CCD |
| Detector resolution | 2048 pixels |
| Image resolution | 128 × 128 pixels |
| Imaging area | 28,672 mm (2048 × 14 µm) |
| Pixel pitch | 14 µm |
| Detector sensitivity | 1800 (V/Lx.S) @ 660 nm |
| Light source | Encapsulated light-emitting diodes (eLEDs) |
| Wavelengths | 2 × (690, 750, 800, and 850 nm) |
| Distance from CCD | 15 mm |
| Max. frame rate | 24 |
| Power consumption | 100 mA @ 5V |
| Radiated power | 20 mA |
| Patient | Tool | Tumor Size (cm) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Pretreatment | Post- Treatment 1 | Post- Treatment 2 | Post- Treatment 3 | Post- Treatment 4 | Post- Treatment 5 | Post- Treatment 6 | Post-Chemo | ||
| 12 | PALP | 2.5 × 2.5 | 3 × 3 | NP | NP | NP | NP | NP | NA |
| US | 3.2 × 1.3 × 2.0 | NA | NA | NA | NA | NA | NA | NA | |
| 13 | PALP | 10 × 9 | 7 × 8 | 5 × 6 | 3.5 × 3.5 | 2.5 × 2 | 2.5 × 2 | NP | NA |
| US | 5.0 × 5.1 × 4.1 | NA | NA | NA | NA | 3.2 × 1.4 × 1.7 | NA | NA | |
| 26 | PALP | 8 × 10 3 | 4 × 3 | NA | 5 × 5.5 | 3 | NA | 5.5 × 5.5 | NA |
| US | 3.8 × 3.9 × 2.3 1.9 × 1.7 × 1.9 | NA | NA | NA | 1.0 × 1.3 × 0.9 1.0 × 1.0 × 0.5 | NA | NA | 0.9 × 1.0 × 0.7 0.4 × 0.4 × 0.5 | |
| Patient | AOC | Pretreatment | Post- Treatment 1 | Post- Treatment 2 | Post- Treatment 3 | Post- Treatment 4 | Post- Treatment 5 | Post- Treatment 6 |
|---|---|---|---|---|---|---|---|---|
| 12 | Unhealthy | 1680.1 | 1669.2 | 1679.3 | 1667.2 | 1649.3 | 1652.1 | NA |
| Healthy | 1657.1 | 1657.1 | 1657.1 | 1657.1 | 1657.1 | 1657.1 | NA | |
| Error | 22.96 | 12.04 | 22.17 | 9.98 | 0.00 | 0.00 | NA | |
| 13 | Unhealthy | 1902.9 | 1929.2 | 1896.1 | 1869.0 | 1844.2 | 1833.2 | 1821.5 |
| Healthy | 1838.4 | 1838.4 | 1838.4 | 1838.4 | 1838.4 | 1838.4 | 1838.4 | |
| Error | 64.56 | 90.88 | 57.77 | 30.63 | 5.89 | 0.00 | 0.00 | |
| 26 | Unhealthy | 1633.6 | 1627.3 | NA | NA | NA | NA | 1627.2 |
| Healthy | 1603.7 | 1593.0 | NA | NA | NA | NA | 1605.7 | |
| Error | 39.97 | 34.34 | NA | NA | NA | NA | 21.46 |
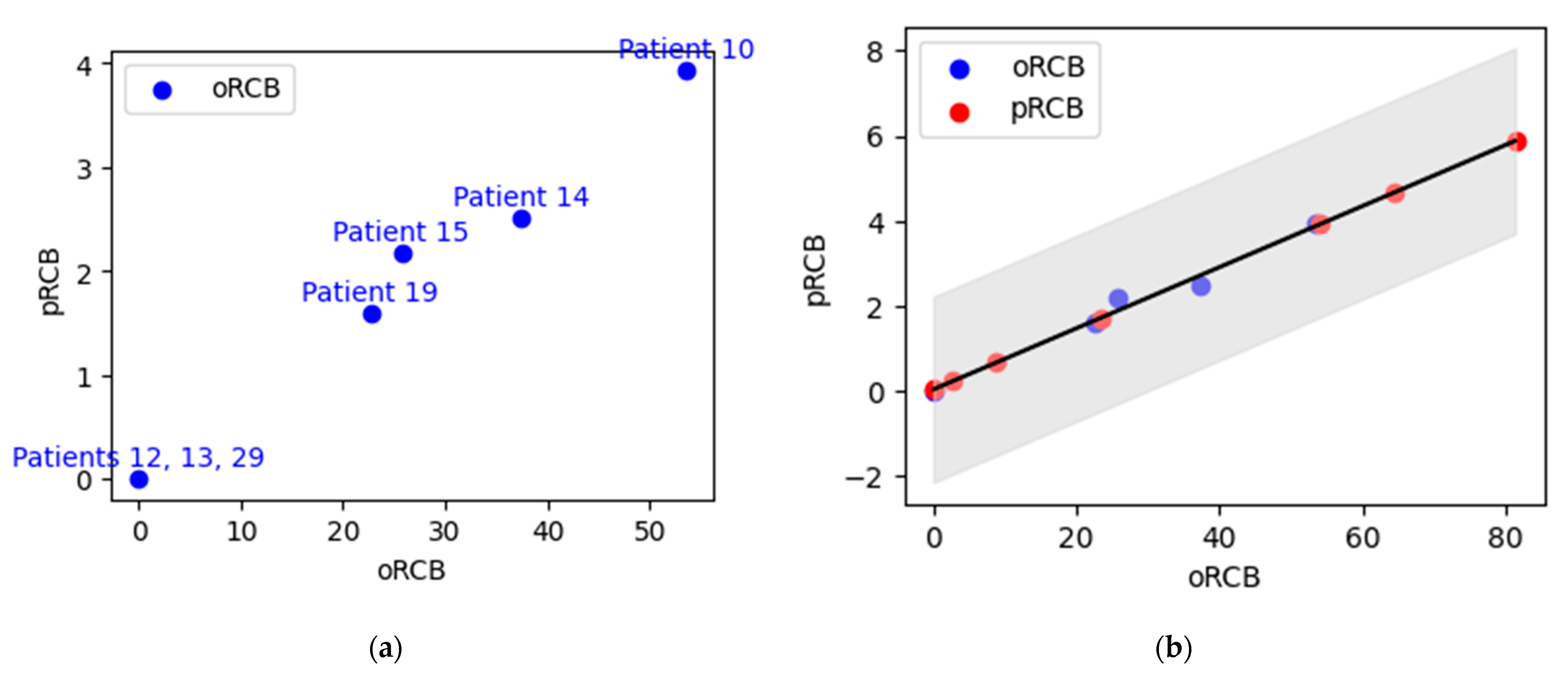
| Patient | Known pRCB Value | Known pRCB Class | Treatment | Unhealthy | Healthy | Error | Predicted oRCB | Predicted Unknown pRCB Value | Predicted Unknown pRCB Class |
|---|---|---|---|---|---|---|---|---|---|
| 10 | 3.93 | RCB-III | Pre-t | 1914.8 | 1839.3 | 75.45 | 53.54 | NA | NA |
| Post-t7 | 1882.9 | 1842.4 | 40.40 | ||||||
| 12 | 0.00 | RCB-0 | Pre-t | 1680.1 | 1657.1 | 22.96 | 0.00 | NA | NA |
| Post-t5 | 1652.1 | 1657.1 | 0.00 | ||||||
| 13 | 0.00 | RCB-0 | Pre-t | 1902.9 | 1838.4 | 64.56 | 0.00 | NA | NA |
| Post-t6 | 1821.5 | 1838.4 | 0 | ||||||
| 14 | 2.51 | RCB-II | Pre-t | 1657.7 | 1614.1 | 43.64 | 37.37 | NA | NA |
| Post-t7 | 1630.4 | 1614.1 | 16.31 | ||||||
| 15 | 2.18 | RCB-II | Pre-t | 1707.1 | 1601.6 | 105.51 | 25.75 | NA | NA |
| Post-t7 | 1630 | 1602.8 | 27.176 | ||||||
| 16 | NA | NA | Pre-t | 2144.2 | 1983.3 | 160.95 | 81.34 | 5.88 | RCB-III |
| Post-t3 | 2122.5 | 1991.6 | 130.92 | ||||||
| 17 | NA | NA | Post-t1 | 2035 | 1981.5 | 53.463 | 8.75 | 0.67 | RCB-I |
| Post-t7 | 1995.6 | 1990.9 | 4.679 | ||||||
| 18 | Lack of Data | ||||||||
| 19 | 1.6 | RCB-II | Pre-t | 1860.5 | 1815.4 | 45.15 | 22.7 | NA | NA |
| Post-t5 | 1825.6 | 1815.4 | 10.25 | ||||||
| 21 | NA | NA | Post-t4 | 1603.5 | 1588.8 | 14.69 | 54.15 | 3.93 | RCB-III |
| Post-t7 | 1615.5 | 1607.6 | 7.96 | ||||||
| 22 | NA | NA | Post-t1 | 2034.5 | 1995.8 | 38.72 | 23.47 | 1.73 | RCB-II |
| Post-t6 | 2014.5 | 2005.4 | 9.09 | ||||||
| 26 | NA | NA | Post-t1 | 1627.3 | 1594.0 | 33.34 | 64.37 | 4.66 | RCB-III |
| Post-t6 | 1627.2 | 1605.7 | 21.46 | ||||||
| 29 | 0.00 | RCB-0 | Post-t2 | 2074.2 | 2073.3 | 0.89 | 0.00 | NA | NA |
| PC | 2028 | 2054.8 | 0.00 | ||||||
| 30 | NA | NA | Pre-t | 2018.2 | 1983.3 | 34.91 | 2.67 | 0.23 | RCB-I |
| Post-t5 | 1976.6 | 1975.7 | 0.93 | ||||||
| 31 | NA | NA | Pre-t | 2025.3 | 1999.3 | 25.99 | 0.00 | 0.00 | RCB-0 |
| Post-t5 | 2001.1 | 2015.3 | 0.00 | ||||||
| Patient | oRCB Value | pRCB Value | Lower Confidence Interval | Upper Confidence Interval |
|---|---|---|---|---|
| 16 | 81.34 | 5.88 | 3.70 | 8.06 |
| 17 | 8.75 | 0.67 | −1.51 | 2.85 |
| 21 | 54.15 | 3.93 | 1.75 | 6.11 |
| 22 | 23.47 | 1.73 | −0.45 | 3.91 |
| 26 | 64.37 | 4.66 | 2.49 | 6.84 |
| 30 | 2.67 | 0.23 | −1.94 | 2.41 |
| 31 | 0.00 | 0.00 | −2.13 | 2.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Momtahen, S.; Momtahen, M.; Ramaseshan, R.; Golnaraghi, F. An Optical Sensory System for Assessment of Residual Cancer Burden in Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy. Sensors 2023, 23, 5761. https://doi.org/10.3390/s23125761
Momtahen S, Momtahen M, Ramaseshan R, Golnaraghi F. An Optical Sensory System for Assessment of Residual Cancer Burden in Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy. Sensors. 2023; 23(12):5761. https://doi.org/10.3390/s23125761
Chicago/Turabian StyleMomtahen, Shadi, Maryam Momtahen, Ramani Ramaseshan, and Farid Golnaraghi. 2023. "An Optical Sensory System for Assessment of Residual Cancer Burden in Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy" Sensors 23, no. 12: 5761. https://doi.org/10.3390/s23125761
APA StyleMomtahen, S., Momtahen, M., Ramaseshan, R., & Golnaraghi, F. (2023). An Optical Sensory System for Assessment of Residual Cancer Burden in Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy. Sensors, 23(12), 5761. https://doi.org/10.3390/s23125761








