A Real-Time Evaluation Algorithm for Noncontact Heart Rate Variability Monitoring
Abstract
:1. Introduction
1.1. Background
1.2. Related Works
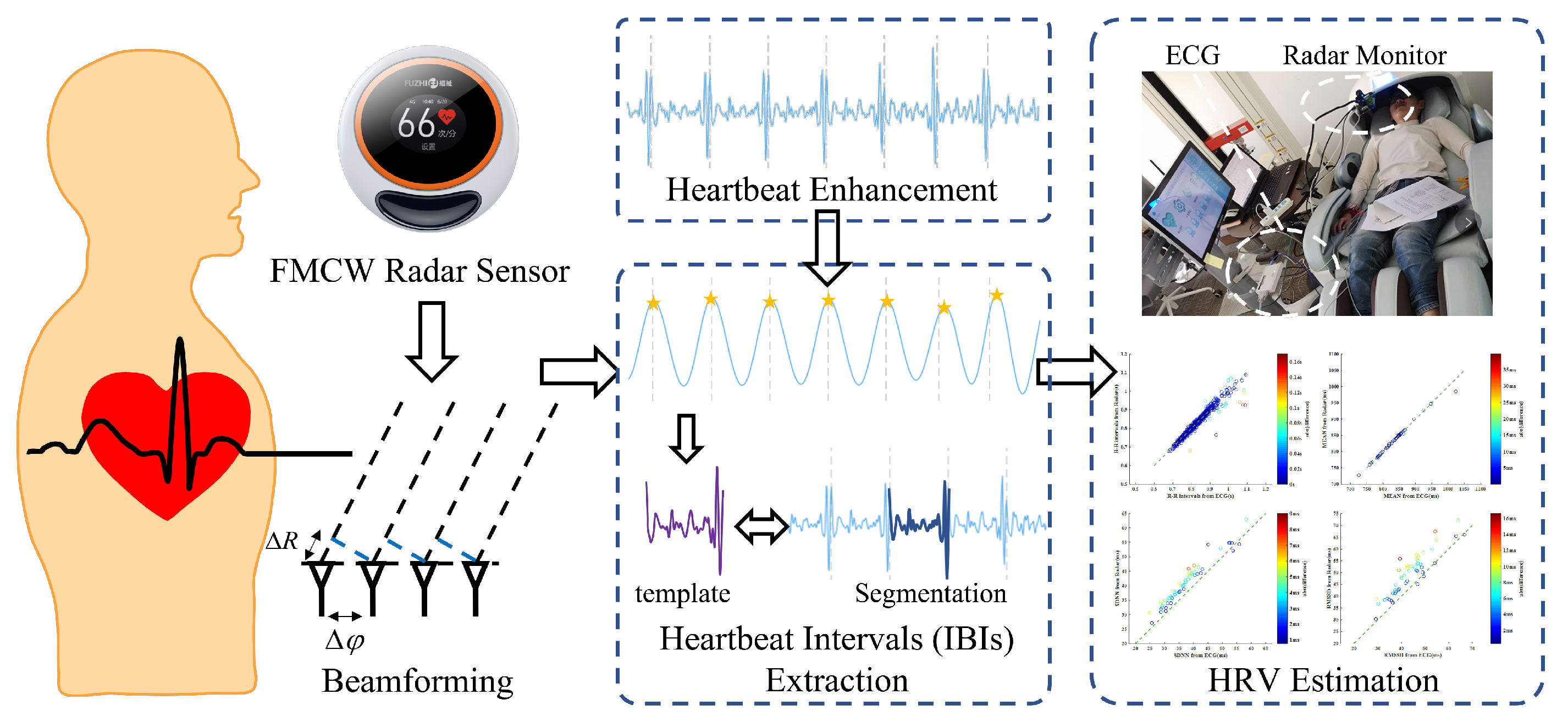
2. Algorithms
2.1. FMCW Radar Signal Preprocessing
2.2. Heartbeat Signal Enhancement
2.3. Accurate Extraction of Heartbeat Intervals
2.4. Algorithm Summary and Convergence Conditions
| Algorithm 1 Heartbeat Interval Extraction |
| Require: The demodulated chest wall motion signal Ensure: The optimal segmentation
|
3. Experiments
4. Results and Discussion
Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HRV | Heart Rate Variability |
| FMCW | Frequency-modulated continuous wave |
| IBI | Interbeat interval |
| RMSE | Root mean square error |
| SDNN | Standard deviation of normal to normal |
| RMSSD | Root mean square of successive differences |
| ANS | Autonomic nervous system |
| PNS | Parasympathetic nervous system |
| SNS | Sympathetic nervous system |
| ECG | Electrocardiography |
| PPG | photoplethysmography |
| OSA | obstructive sleep apnea |
| SIDS | Sudden infant death syndrome |
| CW | Continuous wave |
| EMD | Empirical mode decomposition |
| IMF | Intrinsic mode function |
| DE | Differential enhancement |
| HR | Heartbeat rate |
| MIMO | Multiple input multiple output |
| IF | Intermediate frequency |
| ADC | Analog-to-digital converter |
| FFT | Fast Fourier transform |
| MVDR | Minimum variance distortionless response |
| EVM | Evaluation module |
| Tx | Transmitting antennas |
| Rx | receiving antennas |
| CDF | Cumulative distribution function |
| FTPR | Frequency time phase regression |
| BPF | Bandpass filter |
| FOV | Field of view |
References
- Kunkels, Y.K.; van Roon, A.M.; Wichers, M.; Riese, H. Cross-instrument feasibility, validity, and reproducibility of wireless heart rate monitors: Novel opportunities for extended daily life monitoring. Psychophysiology 2021, 58, e13898. [Google Scholar] [PubMed]
- Venkatesan, C.; Karthigaikumar, P.; Paul, A.; Satheeskumaran, S.; Kumar, R. ECG signal preprocessing and SVM classifier-based abnormality detection in remote healthcare applications. IEEE Access 2018, 6, 9767–9773. [Google Scholar]
- Lehrer, P.; Kaur, K.; Sharma, A.; Shah, K.; Huseby, R.; Bhavsar, J.; Sgobba, P.; Zhang, Y. Heart rate variability biofeedback improves emotional and physical health and performance: A systematic review and meta analysis. Appl. Psychophysiol. Biofeedback 2020, 45, 109–129. [Google Scholar]
- Zhai, Q.; Tang, T.; Lu, X.; Zhou, X.; Li, C.; Yi, J.; Liu, T. Machine Learning-Enabled Noncontact Sleep Structure Prediction. Adv. Intell. Syst. 2022, 4, 2100227. [Google Scholar] [CrossRef]
- Koch, C.; Wilhelm, M.; Salzmann, S.; Rief, W.; Euteneuer, F. A meta-analysis of heart rate variability in major depression. Psychol. Med. 2019, 49, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Ma, L. Depression and cardiovascular disease in elderly: Current understanding. J. Clin. Neurosci. 2018, 47, 1–5. [Google Scholar]
- Lacuesta, R.; Garcia, L.; García-Magariño, I.; Lloret, J. System to recommend the best place to live based on wellness state of the user employing the heart rate variability. IEEE Access 2017, 5, 10594–10604. [Google Scholar]
- Faust, O.; Hong, W.; Loh, H.W.; Xu, S.; Tan, R.S.; Chakraborty, S.; Barua, P.D.; Molinari, F.; Acharya, U.R. Heart rate variability for medical decision support systems: A review. Comput. Biol. Med. 2022, 145, 105407. [Google Scholar] [CrossRef]
- Zhao, F.; Li, M.; Qian, Y.; Tsien, J.Z. Remote measurements of heart and respiration rates for telemedicine. PLoS ONE 2013, 8, e71384. [Google Scholar]
- De Haan, G.; Van Leest, A. Improved motion robustness of remote-PPG by using the blood volume pulse signature. Physiol. Meas. 2014, 35, 1913. [Google Scholar]
- Butler, M.; Crowe, J.; Hayes-Gill, B.; Rodmell, P. Motion limitations of non-contact photoplethysmography due to the optical and topological properties of skin. Physiol. Meas. 2016, 37, N27. [Google Scholar]
- AlNaji, A.; Gibson, K.; Lee, S.H.; Chahl, J. Monitoring of cardiorespiratory signal: Principles of remote measurements and review of methods. IEEE Access 2017, 5, 15776–15790. [Google Scholar] [CrossRef]
- Al-Naji, A.; Gibson, K.; Chahl, J. Remote sensing of physiological signs using a machine vision system. J. Med. Eng. Technol. 2017, 41, 396–405. [Google Scholar]
- Brüser, C.; Antink, C.H.; Wartzek, T.; Walter, M.; Leonhardt, S. Ambient and unobtrusive cardiorespiratory monitoring techniques. IEEE Rev. Biomed. Eng. 2015, 8, 30–43. [Google Scholar] [CrossRef]
- Li, C.; Cummings, J.; Lam, J.; Graves, E.; Wu, W. Radar remote monitoring of vital signs. IEEE Microw. Mag. 2009, 10, 47–56. [Google Scholar]
- Hu, W.; Zhao, Z.; Wang, Y.; Zhang, H.; Lin, F. Noncontact accurate measurement of cardiopulmonary activity using a compact quadrature Doppler radar sensor. IEEE Trans. Biomed. Eng. 2013, 61, 725–735. [Google Scholar]
- Li, C.; Ling, J.; Li, J.; Lin, J. Accurate Doppler radar noncontact vital sign detection using the RELAX algorithm. IEEE Trans. Instrum. Meas. 2009, 59, 687–695. [Google Scholar]
- Zakrzewski, M.; Raittinen, H.; Vanhala, J. Comparison of center estimation algorithms for heart and respiration monitoring with microwave Doppler radar. IEEE Sens. J. 2011, 12, 627–634. [Google Scholar]
- Wang, G.; Gu, C.; Inoue, T.; Li, C. A hybrid FMCW-interferometry radar for indoor precise positioning and versatile life activity monitoring. IEEE Trans. Microw. Theory Tech. 2014, 62, 2812–2822. [Google Scholar]
- Zhai, Q.; Han, X.; Han, Y.; Yi, J.; Wang, S.; Liu, T. A Contactless On-Bed Radar System for Human Respiration Monitoring. IEEE Trans. Instrum. Meas. 2022, 71, 1–10. [Google Scholar]
- Massagram, W.; Lubecke, V.M.; HØst-Madsen, A.; Boric-Lubecke, O. Assessment of heart rate variability and respiratory sinus arrhythmia via Doppler radar. IEEE Trans. Microw. Theory Tech. 2009, 57, 2542–2549. [Google Scholar]
- Hosseini, S.M.A.T.; Amindavar, H. A new Ka-band Doppler radar in robust and precise cardiopulmonary remote sensing. IEEE Trans. Instrum. Meas. 2017, 66, 3012–3022. [Google Scholar] [CrossRef]
- Petrović, V.L.; Janković, M.M.; Lupšić, A.V.; Mihajlović, V.R.; Popović-Božović, J.S. High-accuracy real-time monitoring of heart rate variability using 24 GHz continuous-wave Doppler radar. IEEE Access 2019, 7, 74721–74733. [Google Scholar] [CrossRef]
- Shih, J.Y.; Wang, F.K. Quadrature Cosine Transform (QCT) With Varying Window Length (VWL) Technique for Noncontact Vital Sign Monitoring Using a Continuous-Wave (CW) Radar. IEEE Trans. Microw. Theory Tech. 2021, 70, 1639–1650. [Google Scholar]
- Antolinos, E.; García-Rial, F.; Hernández, C.; Montesano, D.; Godino-Llorente, J.I.; Grajal, J. Cardiopulmonary activity monitoring using millimeter wave radars. Remote Sens. 2020, 12, 2265. [Google Scholar]
- Lee, H.; Kim, B.H.; Park, J.K.; Yook, J.G. A novel vital-sign sensing algorithm for multiple subjects based on 24-GHz FMCW Doppler radar. Remote Sens. 2019, 11, 1237. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.I.; Song, W.J.; Song, H.; Shin, H.C. Improved Heartbeat Detection by Exploiting Temporal Phase Coherency in FMCW Radar. IEEE Access 2021, 9, 163654–163664. [Google Scholar]
- Choi, H.-I.; Song, W.-J.; Song, H.; Shin, H.-C. Selecting target range with accurate vital sign using spatial phase coherency of FMCW radar. Appl. Sci. 2021, 11, 4514. [Google Scholar]
- Choi, H.I.; Song, H.; Shin, H.C. Target range selection of FMCW radar for accurate vital information extraction. IEEE Access 2020, 9, 1261–1270. [Google Scholar]
- Lv, W.; Zhao, Y.; Zhang, W.; Liu, W.; Hu, A.; Miao, J. Remote Measurement of Short-Term Heart Rate with Narrow Beam Millimeter Wave Radar. IEEE Access 2021, 9, 165049–165058. [Google Scholar] [CrossRef]
- Mercuri, M.; Sacco, G.; Hornung, R.; Zhang, P.; Visser, H.J.; Hijdra, M.; Liu, Y.H.; Pisa, S.; Van Liempd, B.; Torfs, T. 2-D localization, angular separation and vital signs monitoring using a SISO FMCW radar for smart long-term health monitoring environments. IEEE Internet Things J. 2021, 8, 11065–11077. [Google Scholar] [CrossRef]
- Wang, Y.; Shui, Y.; Yang, X.; Li, Z.; Wang, W. Multi-target vital signs detection using frequency-modulated continuous wave radar. EURASIP J. Adv. Signal Process. 2021, 2021, 103. [Google Scholar]
- Kim, J.Y.; Park, J.H.; Jang, S.Y.; Yang, J.R. Peak detection algorithm for vital sign detection using Doppler radar sensors. Sensors 2019, 19, 1575. [Google Scholar]
- Nosrati, M.; Tavassolian, N. High-accuracy heart rate variability monitoring using Doppler radar based on Gaussian pulse train modeling and FTPR algorithm. IEEE Trans. Microw. Theory Tech. 2017, 66, 556–567. [Google Scholar]
- Zhang, J.; Wu, Y.; Chen, Y.; Chen, T. Health-radio: Towards contactless myocardial infarction detection using radio signals. IEEE Trans. Mob. Comput. 2020, 21, 585–597. [Google Scholar]
- Sun, L.; Huang, S.; Li, Y.; Gu, C.; Pan, H.; Hong, H.; Zhu, X. Remote measurement of human vital signs based on joint-range adaptive EEMD. IEEE Access 2020, 8, 68514–68524. [Google Scholar]
- Lv, W.; He, W.; Lin, X.; Miao, J. Non-contact monitoring of human vital signs using FMCW millimeter wave radar in the 120 GHz band. Sensors 2021, 21, 2732. [Google Scholar] [PubMed]
- Xiong, Y.; Peng, Z.; Gu, C.; Li, S.; Wang, D.; Zhang, W. Differential enhancement method for robust and accurate heart rate monitoring via microwave vital sign sensing. IEEE Trans. Instrum. Meas. 2020, 69, 7108–7118. [Google Scholar]
- Zhao, M.; Adib, F.; Katabi, D. Emotion Recognition Using Wireless Signals. Commun. ACM 2018, 61, 91–100. [Google Scholar]
- Wen, L.; Gao, Y.; Gu, C.; Mao, J. PhysioChair: A Dual-Frequency Radar System for Noninvasive and Continuous Detection of Physiological Signatures. IEEE Sens. J. 2022, 22, 8224–8233. [Google Scholar]
- Matsui, T.; Katayose, S. A novel method to estimate changes in stress-induced salivary α-amylase using heart rate variability and respiratory rate, as measured in a non-contact manner using a single radar attached to the back of a chair. J. Med. Eng. Technol. 2014, 38, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Konishi, T.; Kawamura, H.; Furuta, S.; Hirobayashi, S.; Yoshida, K. Validation of CW Doppler Radar Embedded Within Driving Seat For Heartbeat Detection Using NHA. In Proceedings of the 2021 IEEE 3rd Global Conference on Life Sciences and Technologies (LifeTech), Nara, Japan, 9–11 March 2021; pp. 317–318. [Google Scholar]
- Berenschot, L. Vital Signs Monitoring with an FMCW Radar Sensor. Bachelor’s Thesis, University of Twente, Enschede, The Netherlands, 2022. [Google Scholar]
- Huang, X.; Ju, Z.; Zhang, R. Real-Time Heart Rate Detection Method Based on 77 GHz FMCW Radar. Micromachines 2022, 13, 1960. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Zhou, W.; Ren, Y.; Wang, J.; Guo, L. Non-Contact Heart Rate Monitoring Based on Millimeter Wave Radar. IEEE Access 2022, 10, 74033–74044. [Google Scholar] [CrossRef]
- Wang, F.; Zeng, X.; Wu, C.; Wang, B.; Liu, K.R. mmhrv: Contactless heart rate variability monitoring using millimeter-wave radio. IEEE Internet Things J. 2021, 8, 16623–16636. [Google Scholar] [CrossRef]











| Subject ID | MEAN (ms) | SDNN (ms) | RMSSD (ms) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Radar | ECG | Err | Radar | ECG | Err | Radar | ECG | Err | |
| 1 | 945.6 | 947.2 | 1.5 (0.16%) | 54.7 | 52.3 | 2.4 (4.65%) | 65.4 | 63.2 | 2.2 (3.46%) |
| 2 | 809.0 | 809.5 | 0.5 (0.06%) | 35.6 | 31.7 | 3.9 (12.3%) | 52.5 | 44.3 | 8.3 (18.7%) |
| 3 | 758.7 | 759.4 | 0.7 (0.09%) | 44.7 | 40.6 | 4.1 (10.2%) | 48.7 | 41.3 | 7.3 (17.8%) |
| 4 | 812.6 | 815.3 | 2.6 (0.33%) | 43.7 | 38.2 | 5.4 (14.2%) | 48.5 | 40.3 | 8.1 (20.2%) |
| 5 | 786.0 | 786.4 | 0.4 (0.05%) | 33.9 | 32.3 | 1.6 (5.00%) | 38.0 | 35.9 | 2.1 (5.96%) |
| 6 | 864.0 | 865.1 | 1.1 (0.12%) | 44.3 | 42.6 | 1.7 (3.94%) | 52.1 | 48.7 | 3.5 (7.11%) |
| 7 | 853.9 | 854.7 | 0.8 (0.09%) | 33.9 | 31.2 | 2.7 (8.81%) | 41.2 | 36.5 | 4.7 (12.8%) |
| 8 | 850.3 | 851.3 | 0.9 (0.11%) | 44.5 | 39.6 | 4.9 (12.3%) | 57.5 | 46.6 | 10.9 (23.5%) |
| 9 | 846.7 | 847.1 | 0.3 (0.04%) | 37.7 | 33.6 | 4.1 (12.2%) | 56.8 | 47.3 | 9.6 (20.2%) |
| 10 | 850.5 | 850.8 | 0.3 (0.04%) | 44.1 | 39.5 | 4.6 (11.7%) | 63.9 | 55.0 | 8.8 (16.1%) |
| Average | 0.91 (0.11%) | 3.5 (9.53%) | 6.6 (14.6%) | ||||||
| Ref. | RF Equipment | Distance (m) | HR ACC. (%) | HRV Monitoring | IBI RMSE (ms) |
|---|---|---|---|---|---|
| [43] | AWR1642 (TI) | 0.5 | ⩾90% | No | - |
| [44] | IWR1443 (TI) | 0.28–0.7 | 97% | No | - |
| [45] | IWR1843 (TI) | 0.5–2.0 | 98% | No | - |
| [46] | IWR1843 (TI) | 0.5 | Not Mentioned | Yes | 26.06 |
| Ours | IWR1843 (TI) | 0.5 | ⩾98% | Yes | 14.9 |
| The Proportion of Measurements with a Measurement Error of | MEAN | SDNN | RMSSD | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BPF | DE | Ours | BPF | DE | Ours | BPF | DE | Ours | |
| ⩽5 ms | 15.0% | 72.5% | 90.0% | 7.5% | 35.0% | 70.0% | 2.5% | 12.5% | 37.5% |
| ⩽10 ms | 27.5% | 95.0% | 97.5% | 10.0% | 50.0% | 97.5% | 2.5% | 27.5% | 80.0% |
| ⩽15 ms | 32.5% | 100% | 100% | 15.0% | 65.0% | 100% | 12.5% | 45.0% | 87.5% |
| ⩽20 ms | 42.5% | - | - | 22.5% | 70.0% | - | 20.0% | 50.0% | 95.0% |
| ⩽30 ms | 65.0% | - | - | 40.0% | 95.0% | - | 27.5% | 65.0% | 100% |
| ⩽40 ms | 70.0% | - | - | 50.0% | 97.5% | - | 30.0% | 75.0% | - |
| ⩽50 ms | 77.5% | - | - | 52.5% | 100% | - | 37.5% | 87.5% | - |
| RMSE (ms) | 46.30 | 4.92 | 3.24 | 88.21 | 16.36 | 4.91 | 117.70 | 31.54 | 9.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, X.; Zhai, Q.; Zhang, N.; Zhang, X.; He, L.; Pan, M.; Zhang, B.; Liu, T. A Real-Time Evaluation Algorithm for Noncontact Heart Rate Variability Monitoring. Sensors 2023, 23, 6681. https://doi.org/10.3390/s23156681
Han X, Zhai Q, Zhang N, Zhang X, He L, Pan M, Zhang B, Liu T. A Real-Time Evaluation Algorithm for Noncontact Heart Rate Variability Monitoring. Sensors. 2023; 23(15):6681. https://doi.org/10.3390/s23156681
Chicago/Turabian StyleHan, Xiangyu, Qian Zhai, Ning Zhang, Xiufeng Zhang, Long He, Min Pan, Bin Zhang, and Tao Liu. 2023. "A Real-Time Evaluation Algorithm for Noncontact Heart Rate Variability Monitoring" Sensors 23, no. 15: 6681. https://doi.org/10.3390/s23156681
APA StyleHan, X., Zhai, Q., Zhang, N., Zhang, X., He, L., Pan, M., Zhang, B., & Liu, T. (2023). A Real-Time Evaluation Algorithm for Noncontact Heart Rate Variability Monitoring. Sensors, 23(15), 6681. https://doi.org/10.3390/s23156681







