3.1. Basecaller Comparison
The experiments were conducted on three publicly available datasets on
Acinetobacter pittii,
Haemophilus haemolyticus, and
Serratia marcescens genomes, sequenced using MinION Flow Cell (R9.4.1) (Oxford Nanopore Technologies, Oxford, United Kingdom). The datasets were downloaded as raw fast5 files from the [
18] study, composed of 4467, 8670, and 16,743 files for
Acinetobacter pittii,
Haemophilus haemolyticus, and
Serratia marcescens, respectively. The downloaded data were basecalled by Albacore (ver. 3.0.1), Causalcall (commit f5ab3db), Chiron (ver. 0.6.1.1), Dorado [
25] (ver. 0.2.4), Flappie (ver. 2.1.3), Guppy (ver. 3.3.0), and Nanonet (ver. 2.0.0). The launch parameters for all applications are posted in the publicly available repository:
https://github.com/wkusmirek/basecalling-quality (accessed on 13 June 2023). To explore the effect of the training set on the estimation reconstruction quality performance, for the Dorado basecaller, we used three trained, default models (
[email protected],
[email protected], and
[email protected]), and for the Guppy tool, we used the default model and two custom trained models downloaded from the [
18] study (the Guppy tools with custom models are marked in the presented paper as ‘Guppy kp’ and ‘Guppy kp-big-net’). Briefly, the models were trained by the Sloika neural network training toolkit (
https://github.com/nanoporetech/sloika (accessed on 13 June 2023)) on the training dataset composed of reads from 50 different isolate genomes. The final training dataset contained 226,166 DNA reads for training the neural network and 1000 DNA reads for validating the results. A detailed description of the model’s training process and the scripts used can be found in reference [
18].
As a result of the basecalling process, we obtained 33 sets of long DNA reads; the basic statistics of the obtained reads calculated with the BBMap [
26] tool are presented in
Table 2. Firstly, the number of reads basecalled by the Albacore, Causalcall, Chiron, Dorado, Flappie, and Guppy tools were equal to the number of raw fast5 files obtained from the nanopore sequencer. However, the Nanonet tool produced almost twice as many DNA sequences: 7702 reads from 4467 raw fast5 files (
Acinetobacter pittii), 15,956 reads from 8669 raw fast5 files (
Haemophilus haemolyticus), and 34,046 reads from 16,742 raw fast5 files (
Serratia marcescens). Moreover, the sum of DNA reads basecalled by the Nanonet tool was approximately equal to the sum of sequence lengths produced by the other tools, e.g., Albacore. The average length of the basecalled DNA reads we obtained from the Nanonet tool was approximately half the average length of the DNA reads from the other tools, like Albacore. The poor performance of the Nanonet application was also demonstrated in [
18], which confirms that the tool should not be selected for target basecalling in real nanopore DNA sequencing projects. Secondly, the Causalcall, Chiron, and Nanonet tools basecalled DNA reads, which, depending on the dataset, achieved a different mapping factor to the reference genome. For example, the DNA reads basecalled by the Chiron tool were mapped in 81.88%, 43.05%, and 84.50% of the reference genomes for
Acinetobacter pittii,
Haemophilus haemolyticus, and
Serratia marcescens, respectively. Thirdly, the basecalled DNA reads by the Causalcall, Chiron, and Nanonet tools, mapped to the reference genome, were characterized by lower matched rates than the Albacore, Flappie, and Guppy results. Moreover, the Dorado tool with the ‘sup’ model produced DNA reads with the fewest substitution errors among other basecallers for all three investigated datasets.
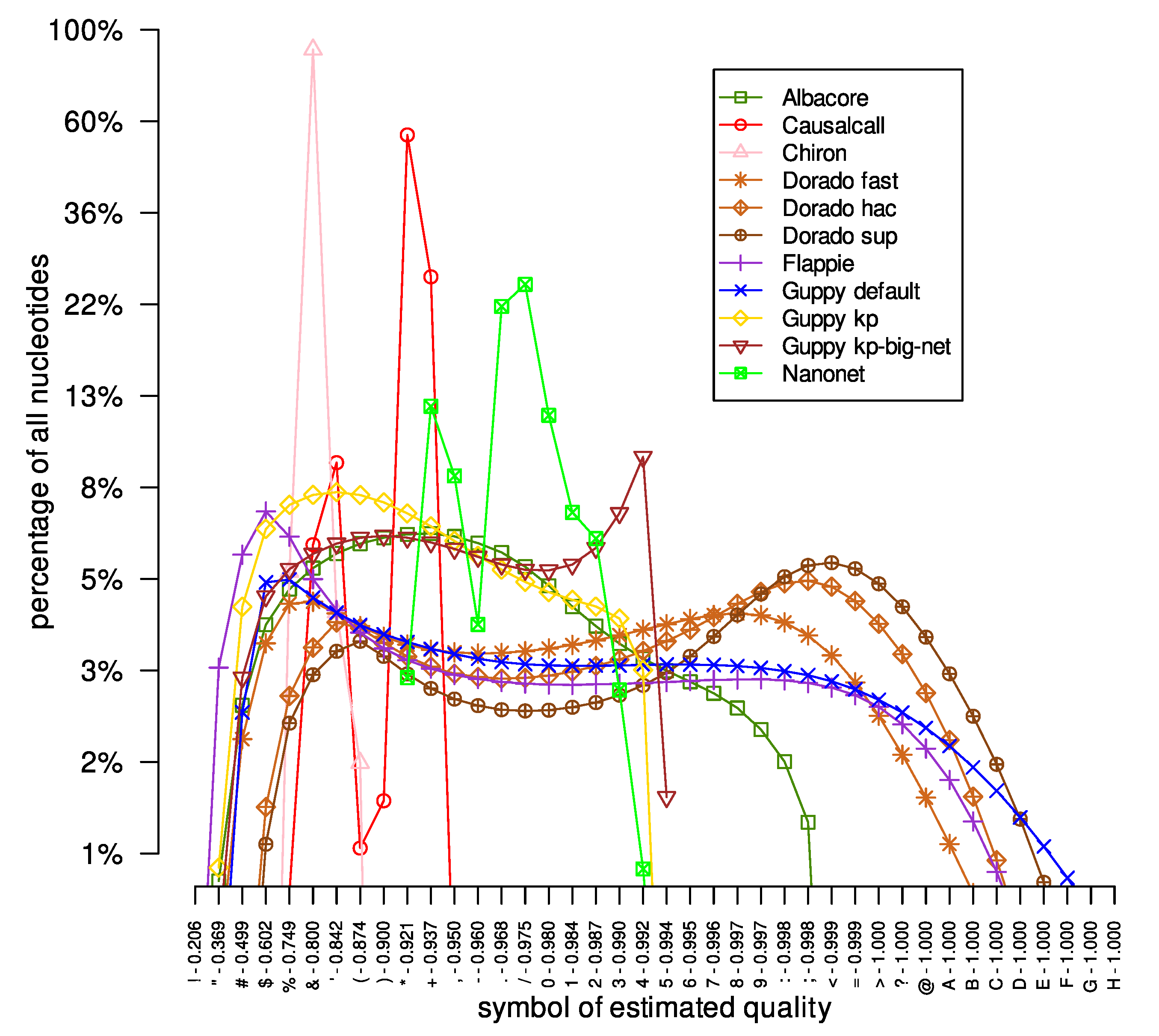
We also checked the distribution of the estimated quality symbols in the investigated DNA reads datasets obtained by different basecallers. The results are presented in
Figure 1. Firstly, different basecallers provide estimated quality symbols from different ranges. For example, the symbols provided by the Flappie application were in the range of ‘"’ to ‘B’, while the Nanonet tool basecalled DNA sequences with quality signs between ‘*’ and ‘3’. Moreover, the same tool with different models outputted nucleotides with estimated quality symbols of a different range. For example, the Guppy tool provides nucleotides with quality symbols ‘#’-‘E’, ‘#’-‘4’, and ‘#’-‘5’ for ‘default’, ‘kp’, and ‘kp-big-net’ models, respectively. Moreover, a different range of quality symbols affects the maximum number of a given quality symbol. For example, for the Albacore tool, the most common quality symbol (‘*’) represents 7% of all of Albacore’s quality symbols. On the other hand, the quality sign ‘&’ represents about 90% of all quality symbols provided by the Chiron tool. The reason for such a large difference in the number of the most common quality symbols is the breadth of the range of quality symbols provided: 4 and 25 other quality signs in the Chiron and Albacore results, respectively. Secondly, the curves representing the distribution of quality symbols for the Causalcall, Chiron, and Nanonet tools are irregular, with large variations in the count values for successive quality symbols, e.g., changes in tens of percents. On the other hand, for the Flappie and Dorado tools, the curves are much smoother, without significant changes in abundance for successive quality symbols. Thirdly, the curves representing the distribution of quality symbols in the results provided by individual basecallers are very similar in all analyzed datasets (see
Supplementary Materials, Figures S1 and S2). For example, the curve of the Flappie application grows rapidly at the beginning, reaching its peak around the ‘&’ and ‘%’ signs, before slowly descending (the same descent curve shape for all three datasets) to the ‘A’ and ‘B’ signs.
After analyzing basic statistics of basecalled DNA reads, we prepared reference genomes by masking repetitive regions in raw reference sequences downloaded from the [
18] study. As a result of this process, we masked 89,222 bp out of 3,814,719 bp (2.34%), 13,860 bp out of 2,042,591 bp (0.68%), and 241,685 bp out of 5,517,578 bp (4.38%) for
Acinetobacter pittii,
Haemophilus haemolyticus, and
Serratia marcescens reference genomes, respectively.
Then, we mapped the long DNA reads onto the previously masked reference genomes. After mapping, for each symbol of estimated quality, we calculated the ratio of the number of mapped nucleotides (nucleotides reported in the SAM file) to the number of all nucleotides with the specified quality sign (nucleotides reported in the FASTQ file). The results are presented in
Figure 2. As expected, with the successive symbols of estimated quality, the ratio of mapped to total nucleotides increases for the results obtained from the Albacore, Dorado, Flappie, and Guppy tools. However, for the DNA reads basecalled by the Causalcall, Chiron, and Nanonet tools, there are situations where even nucleotides with quality symbols denoting more certain probabilities of correct reconstruction were mapped to the reference genome in smaller numbers. Moreover, for the Nanonet application, there are quality symbols that appear in the DNA reads, but not in the mapping results (nucleotides with a given quality symbol do not map to the reference genome). For example, for the
Acinetobacter pittii dataset, the Nanonet results in basecalled reads with estimated quality symbols from symbols ‘*’ to ‘3’. At the same time, the SAM file contains only nucleotides with an estimated reconstruction quality ranging from ‘.’ to ‘3’.
Finally, we computed statistics from each SAM file obtained. The results for the
Acinetobacter pittii dataset are presented in
Figure 3. Briefly, results obtained from the Albacore, Flappie, and Guppy applications had similar characteristics, as expected; the number of matched nucleotides increased with the successive symbols of the estimated quality, and the number of mismatched, inserted, and clipped nucleotides decreased. However, the performance characteristics of the Causalcall, Chiron, and Nanonet applications are irregular. For example, the number of improperly inserted nucleotides for the Nanonet tool results for all three investigated datasets increases with the successive symbols of quality. It is also worth following the course of the curves for the Dorado application; depending on the model used (‘fast’, ‘hac’, or ‘sup’), the curves are different.
3.3. Impact of the Estimated Quality Upon Further Analysis
In the third part of the research presented, we examined the impact of estimated reconstruction quality indicators on the further analysis of genetic data. In particular, we investigated how the estimated quality is used in processes like:
De novo assembling of long DNA reads (Canu [
28], miniasm [
29]);
Resolving ambiguities in the de Bruijn graph (ABySS [
30], SPAdes [
31]);
Linking the results of de novo assembling of short reads by long reads (dnaasm-link [
32], LINKS [
33], SSPACE-LongRead [
34]);
Correcting errors in long reads (Canu [
28], LoRDEC [
35]);
Detecting structural variations (Sniffles [
36]).
In all of the above-mentioned processes, the impact of the estimated quality of nucleotide reconstruction on the results was examined by comparing the results obtained from three datasets: (I) the raw FASTQ file obtained from the Albacore tool with estimated quality symbols, (II) the FASTQ file with fake quality signs—all quality symbols were set to ‘?’ signs, and (III) the FASTA file—the file obtained from the FASTQ file by removing all lines describing estimated quality indicators. It is worth mentioning that the three datasets mentioned contained identical DNA sequences; they differed only in the estimated quality symbols: (I) real quality symbols from the basecaller, (II) the same symbol of estimated quality equal to ‘?’ for all nucleotides, and (III) no reconstruction quality symbols.
All conducted experiments were performed by launching individual applications with the parameters described in the GitHub repository:
https://github.com/wkusmirek/basecalling-quality (accessed on 13 June 2023). The experiments performed, algorithm analysis, and results obtained are described below.
First, we checked how the estimated quality of nucleotide reconstruction in the basecalling process affects de novo assembling results. In the study, we used the Canu [
28] and miniasm [
29] tools. We used three sets (‘FASTQ’, ‘fake FASTQ’, and ‘FASTA’) of DNA reads obtained from
Acinetobacter pittii reference genome. The results were evaluated by the QUAST [
37] tool, the statistics obtained are presented in
Table 4. Briefly, the results obtained with the miniasm application are significantly inferior to those obtained with the Canu software (both applications reconstruct a single scaffold, but the quality of the miniasm resultant DNA sequence is poor). However, both the Canu and miniasm tools allowed obtaining identical results for all three analyzed sets of long DNA reads (‘FASTQ’, ‘fake FASTQ’, and ‘FASTA’), which proves that the symbols of the estimated reconstruction quality do not affect the de novo assembling process of long DNA reads by the Canu and miniasm applications.
Secondly, we examined how the estimated quality of the reconstructed nucleotide can affect the process of resolving ambiguities in the de Bruijn graph [
38]. The short DNA reads of the de novo assembling process usually consist of building the de Bruijn graph and generating the resulting DNA sequences from this structure. One of the main challenges in the de novo assembling process is the presence of repetitive DNA fragments leading to the formation of ambiguous fragments in the graph. Each of the ambiguities in the de Bruijn graph results in shorter resultant DNA sequences, the ambiguities could be resolved by mapping long DNA reads to the de Bruijn graph.
In the study, we used two applications supporting hybrid de novo assembling: the ABySS [
30] and the SPAdes [
31] tools. The experiment was carried out on artificially generated short DNA reads from the
Acinetobacter pittii reference genome using the pIRS application and three sets of long DNA reads (‘FASTQ’, ‘fake FASTQ’, and ‘FASTA’). The resultant sequences were evaluated by the QUAST tool; the obtained statistics are presented in
Table 5. Briefly, the results show that for ABySS and SPAdes, the estimated nucleotide reconstruction quality symbols do not matter in the resolving ambiguities in the de Bruijn graph process; the results for the ‘PET + ONT FASTQ’, ‘PET + ONT fake FASTQ’, and ‘PET + ONT FASTA’ sets are identical. It is also worth noting that for both applications, the usage of long DNA reads improved de novo assembly results in relation to the results obtained only from short DNA reads (rows marked as ‘PET’).
Thirdly, long DNA reads can be used in the scaffolding process—to link de novo assembling results of short DNA reads by long DNA reads. To investigate the impact of the estimated quality of nucleotide reconstruction on the results of combining the resulting DNA sequences of the de novo assembly of short DNA reads by long DNA reads, we used dnaasm-link [
32], LINKS [
33], and SSPACE-LongRead [
34] tools. In the experiment, first, we generated a set of short DNA reads from the
Acinetobacter pittii reference genome by the pIRS [
39] read simulator. Then, the short DNA reads were assembled de novo by the ABySS [
30] tool, the obtained results were scaffolded with the long DNA reads (‘FASTQ’, ‘fake FASTQ’, and ‘FASTA’) by dnaasm-link, LINKS, and SSPACE-LongRead tools. The final scaffolds were evaluated by the QUAST [
37] tool; the results are presented in
Table 6. Briefly, all of the tools linked the contigs by the long DNA read the same way, regardless of the presence and values of estimated nucleotide reconstruction quality symbols.
Fourthly, we checked the impact of the estimated quality of nucleotide reconstruction in the basecalling process on correcting errors in long DNA reads. Correction of long DNA reads can occur in two ways: (I) correcting by short DNA reads, and (II) correcting by other long DNA reads. In the study presented, we used the LoRDEC [
35] and the Canu [
28] tools for correcting long reads by the short DNA reads and other long DNA reads, respectively. In the experiment, we corrected three sets (‘FASTQ’, ‘fake FASTQ’, and ‘FASTA’) of DNA reads obtained from the
Acinetobacter pittii reference genome. The set of short DNA reads was generated from the
Acinetobacter pittii reference genome by the pIRS [
39] reads simulator, and all long DNA reads were evaluated with the BBMap [
26] application; the correction results are presented in
Table 7. Briefly, both the LoRDEC and Canu applications obtained the same correction results for all three datasets (‘ONT FASTQ’, ‘ONT fake FASTQ’, and ‘ONT FASTA’). The estimated nucleotide reconstruction quality symbols are irrelevant in correcting long DNA reads.
Lastly, we checked how the estimated quality determined in the basecalling process affects the number of detected structural variations. In the study, we used the long DNA reads (rel3 release) obtained from the well-characterized NA12787 Genome in a Bottle (GIAB) sample [
40,
41,
42,
43]. The experiment carried out involved (I) mapping long DNA reads to the reference genome by the minimap2 [
24] tool and (II) detecting the structural variations with the Sniffles [
36] application. To speed up the calculations, we limited the research only to chromosome 11; the obtained results are presented in
Table 8. Briefly, the number of structural variations detected is the same for all three sets of long DNA reads. Additionally, the detected changes are identical for all three datasets, e.g., the coordinates of the start and end of the structural variation. In the study, we used only chromosome 11, but the conclusions obtained will be the same for other parts of the genome.