Triple Reporter Assay: A Non-Overlapping Luciferase Assay for the Measurement of Complex Macromolecular Regulation in Cancer Cells Using a New Mushroom Luciferase–Luciferin Pair
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plasmids and Cloning
2.3. Cell Lines, Cell Culture and Transfection
2.4. Protein Sample Preparation and Immunoblotting
2.5. Quantitative RT-PCR
2.6. Live Cell Imaging
2.7. Bioluminescence Imaging (BLI) of Tumor Xenografts
2.8. Chromatin Immunoprecipitation
2.9. Statistical Analysis
3. Results
3.1. Human Codon Optimization Improved Wild-Type Mushroom Luciferase (Luz) Reporter Activity
3.2. Luz and 3OH-Hispidin Combination Is a Novel Non-Cross-Reacting Bioluminescence Reporter System
3.3. NF-κB Regulatory Response Can Be Measured by Using hLuz Reporter Sensor
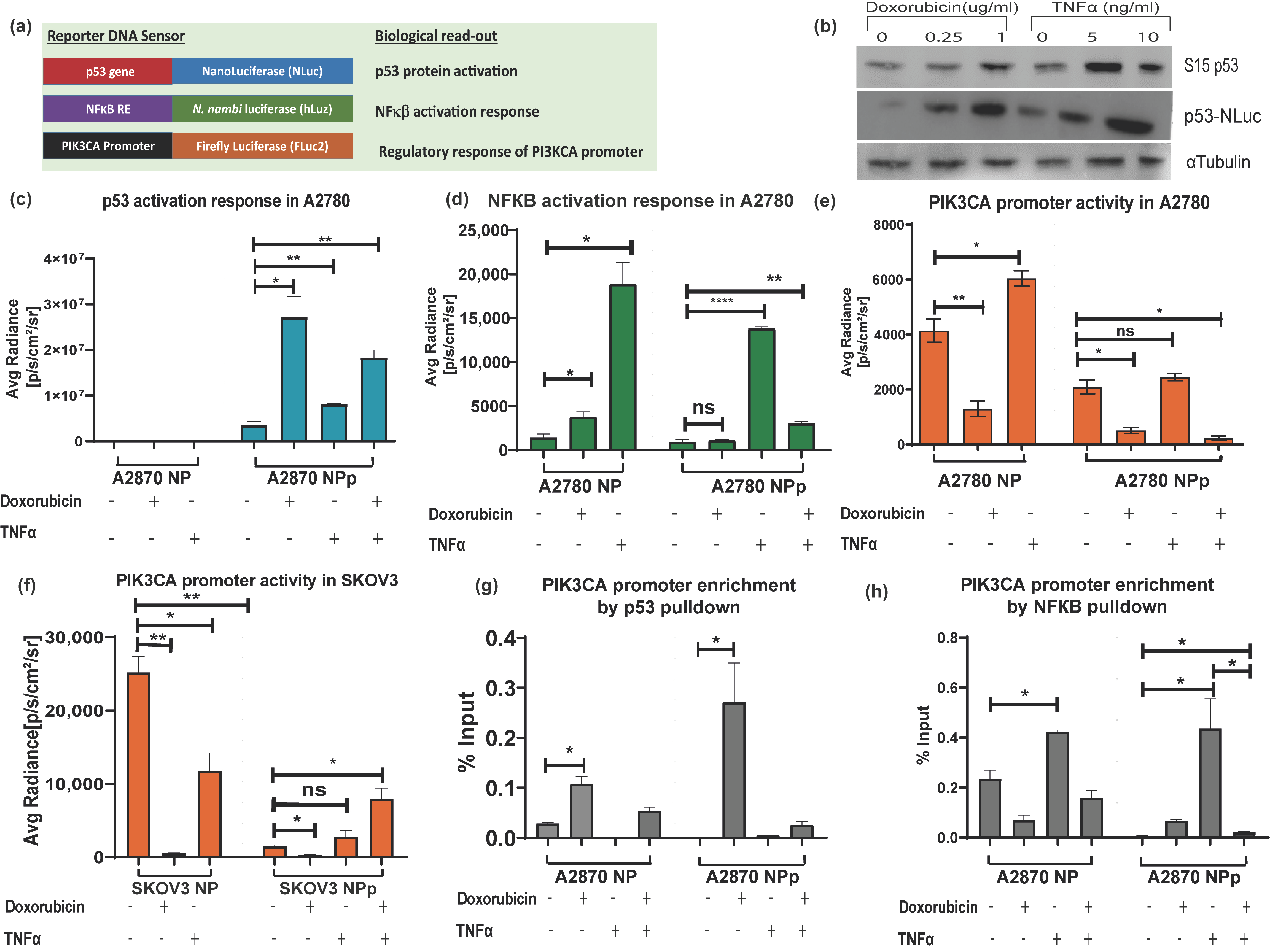
3.4. Application of Triple Luciferase Assay Revealed Complex Regulation of PIK3CA Gene Promoter by p53 and NF-κB
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De, A. Bioluminescence. In eLS; John Wiley & Sons, Ltd: Chichester, UK, 2014. [Google Scholar]
- Kaskova, Z.M.; Tsarkova, A.S.; Yampolsky, I.V. 1001 Lights: Luciferins, Luciferases, Their Mechanisms of Action and Applications in Chemical Analysis, Biology and Medicine. Chem. Soc. Rev. 2016, 45, 6048–6077. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, O.; Johnson, F.H.; Saiga, Y. Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from the Luminous Hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962, 59, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Bhaumik, S.; Gambhir, S.S. Optical Imaging of Renilla Luciferase Reporter Gene Expression in Living Mice. Proc. Natl. Acad. Sci. USA 2002, 99, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Ford, S.R.; Chenault, K.H.; Bunton, L.S.; Hampton, G.J.; McCarthy, J.; Hall, M.S.; Pangburn, S.J.; Buck, L.M.; Leach, F.R. Use of Firefly Luciferase for ATP Measurement: Other Nucleotides Enhance Turnover. J. Biolumin. Chemilumin. 1996, 11, 149–167. [Google Scholar] [CrossRef]
- Li, S.; Ruan, Z.; Zhang, H.; Xu, H. Recent Achievements of Bioluminescence Imaging Based on Firefly Luciferin-Luciferase System. Eur. J. Med. Chem. 2021, 211, 113111. [Google Scholar] [CrossRef]
- Inouye, S.; Shimomura, O. The Use of Renilla Luciferase, Oplophorus Luciferase, and Apoaequorin as Bioluminescent Reporter Protein in the Presence of Coelenterazine Analogues as Substrate. Biochem. Biophys. Res. Commun. 1997, 233, 349–353. [Google Scholar] [CrossRef]
- Kim, S.-B.; Paulmurugan, R. Bioluminescent Imaging Systems for Assay Developments. Anal. Sci. 2021, 37, 233–247. [Google Scholar] [CrossRef]
- Dou, L.; Matz, E.L.; Gu, X.; Shu, F.; Paxton, J.; Song, J.; Yoo, J.; Atala, A.; Jackson, J.; Zhang, Y. Non-Invasive Cell Tracking with Brighter and Red-Transferred Luciferase for Potential Application in Stem Cell Therapy. Cell Transplant. 2019, 28, 1542–1551. [Google Scholar] [CrossRef]
- Guo, Z.; Parakra, R.D.; Xiong, Y.; Johnston, W.A.; Walden, P.; Edwardraja, S.; Moradi, S.V.; Ungerer, J.P.J.; Ai, H.; Phillips, J.J.; et al. Engineering and Exploiting Synthetic Allostery of NanoLuc Luciferase. Nat. Commun. 2022, 13, 789. [Google Scholar] [CrossRef]
- Liu, S.; Su, Y.; Lin, M.Z.; Ronald, J.A. Brightening up Biology: Advances in Luciferase Systems for in Vivo Imaging. ACS Chem. Biol. 2021, 16, 2707–2718. [Google Scholar] [CrossRef]
- Azad, T.; Janse van Rensburg, H.J.; Morgan, J.; Rezaei, R.; Crupi, M.J.F.; Chen, R.; Ghahremani, M.; Jamalkhah, M.; Forbes, N.; Ilkow, C.; et al. Luciferase-Based Biosensors in the Era of the COVID-19 Pandemic. ACS Nanosci. Au 2021, 1, 15–37. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-B.; Fujii, R.; Paulmurugan, R. Ligand-Activatable BRET9 Probes for Imaging Molecular Events in Living Mammalian Cells. In Live Cell Imaging: Methods and Protocols; Springer: New York, NY, USA, 2021; pp. 261–270. [Google Scholar]
- Lopreside, A.; Calabretta, M.M.; Montali, L.; Zangheri, M.; Guardigli, M.; Mirasoli, M.; Michelini, E. Bioluminescence Goes Portable: Recent Advances in Whole-cell and Cell-free Bioluminescence Biosensors. Luminescence 2021, 36, 278–293. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Kim, K.T.; Lindberg, E.; Winssinger, N. Smartphone DNA or RNA Sensing Using Semisynthetic Luciferase-Based Logic Device. ACS Sens. 2020, 5, 807–813. [Google Scholar] [CrossRef]
- Lopreside, A.; Montali, L.; Wang, B.; Tassoni, A.; Ferri, M.; Calabretta, M.M.; Michelini, E. Orthogonal Paper Biosensor for Mercury(II) Combining Bioluminescence and Colorimetric Smartphone Detection. Biosens. Bioelectron. 2021, 194, 113569. [Google Scholar] [CrossRef]
- Hawkins, E.; Butler, B.; Wood, K.; O’grady, M.; Orr, L.; Beck, M.; Dual-GLOTM Luciferase Assay System: A Homogeneous Dual-Reporter System. Promega Notes Mag. 2002. Number 81. Available online: http://www.ulab360.com/files/prod/references/201401/06/541726004.pdf (accessed on 26 July 2023).
- Mcnabb, D.S.; Reed, R.; Marciniak, R.A. Dual Luciferase Assay System for Rapid Assessment of Gene Expression in Saccharomyces Cerevisiae. Eukaryot. Cell 2005, 4, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Sarrion-Perdigones, A.; Chang, L.; Gonzalez, Y.; Gallego-Flores, T.; Young, D.W.; Venken, K.J.T. Examining Multiple Cellular Pathways at Once Using Multiplex Hextuple Luciferase Assaying. Nat. Commun. 2019, 10, 5710. [Google Scholar] [CrossRef]
- Gammon, S.T.; Leevy, W.M.; Gross, S.; Gokel, G.W.; Piwnica-Worms, D. Spectral Unmixing of Multicolored Bioluminescence Emitted from Heterogeneous Biological Sources. Anal. Chem. 2006, 78, 1520–1527. [Google Scholar] [CrossRef]
- Stacer, A.C.; Nyati, S.; Moudgil, P.; Iyengar, R.; Luker, K.E.; Rehemtulla, A.; Luker, G.D. NanoLuc Reporter for Dual Luciferase Imaging in Living Animals. Mol. Imaging 2013, 12, 1–13. [Google Scholar] [CrossRef]
- Purtov, K.V.; Petushkov, V.N.; Baranov, M.S.; Mineev, K.S.; Rodionova, N.S.; Kaskova, Z.M.; Tsarkova, A.S.; Petunin, A.I.; Bondar, V.S.; Rodicheva, E.K.; et al. The Chemical Basis of Fungal Bioluminescence. Angew. Chem.-Int. Ed. 2015, 54, 8124–8128. [Google Scholar] [CrossRef]
- Kaskova, Z.M.; Dörr, F.A.; Petushkov, V.N.; Purtov, K.V.; Tsarkova, A.S.; Rodionova, N.S.; Mineev, K.S.; Guglya, E.B.; Kotlobay, A.; Baleeva, N.S.; et al. Mechanism and Color Modulation of Fungal Bioluminescence. Sci. Adv. 2017, 3, e1602847. [Google Scholar] [CrossRef]
- Teranishi, K. Bioluminescence and Chemiluminescence Abilities of Trans -3-hydroxyhispidin on the Luminous Fungus Mycena Chlorophos. Luminescence 2018, 33, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, M.-J.; Shim, D.; Ryoo, R. De Novo Genome Assembly of the Bioluminescent Mushroom Omphalotus Guepiniiformis Reveals an Omphalotus-Specific Lineage of the Luciferase Gene Block. Genomics 2022, 114, 110514. [Google Scholar] [CrossRef] [PubMed]
- House, C.D.; Jordan, E.; Hernandez, L.; Ozaki, M.; James, J.M.; Kim, M.; Kruhlak, M.J.; Batchelor, E.; Elloumi, F.; Cam, M.C.; et al. NFkB Promotes Ovarian Tumorigenesis via Classical Pathways That Support Proliferative Cancer Cells and Alternative Pathways That Support ALDHþ Cancer Stem–like Cells. Cancer Res. 2017, 77, 6927–6940. [Google Scholar] [CrossRef] [PubMed]
- Harrington, B.S.; Annunziata, C.M. NF-ΚB Signaling in Ovarian Cancer. Cancers 2019, 11, 1182. [Google Scholar] [CrossRef]
- Corney, D.C.; Flesken-Nikitin, A.; Choi, J.; Nikitin, A.Y. Role of P53 and Rb in Ovarian Cancer. In Ovarian Cancer; Springer New York: New York, NY, USA, 2008; pp. 99–117. [Google Scholar]
- Tuna, M.; Ju, Z.; Yoshihara, K.; Amos, C.I.; Tanyi, J.L.; Mills, G.B. Clinical Relevance of TP53 Hotspot Mutations in High-Grade Serous Ovarian Cancers. Br. J. Cancer 2020, 122, 405–412. [Google Scholar] [CrossRef]
- Cheaib, B.; Auguste, A.; Leary, A. The PI3K/Akt/MTOR Pathway in Ovarian Cancer: Therapeutic Opportunities and Challenges. Chin. J. Cancer 2015, 34, 4–16. [Google Scholar] [CrossRef]
- Gasparri, M.; Bardhi, E.; Ruscito, I.; Papadia, A.; Farooqi, A.; Marchetti, C.; Bogani, G.; Ceccacci, I.; Mueller, M.; Benedetti Panici, P. PI3K/AKT/MTOR Pathway in Ovarian Cancer Treatment: Are We on the Right Track? Geburtshilfe Frauenheilkd. 2017, 77, 1095–1103. [Google Scholar] [CrossRef]
- Webster, G.A.; Perkins, N.D. Transcriptional Cross Talk between NF-KappaB and P53. Mol. Cell. Biol. 1999, 19, 3485–3495. [Google Scholar] [CrossRef]
- Gaikwad, S.M.; Gunjal, L.; Junutula, A.R.; Astanehe, A.; Gambhir, S.S.; Ray, P. Non-Invasive Imaging of Phosphoinositide-3-Kinase-Catalytic-Subunit-Alpha (PIK3CA) Promoter Modulation in Small Animal Models. PLoS ONE 2013, 8, e55971. [Google Scholar] [CrossRef]
- Yang, N.; Huang, J.; Greshock, J.; Liang, S.; Barchetti, A.; Hasegawa, K.; Kim, S.; Giannakakis, A.; Li, C.; O’Brien-Jenkins, A.; et al. Transcriptional Regulation of PIK3CA Oncogene by NF-KB in Ovarian Cancer Microenvironment. PLoS ONE 2008, 3, 1758. [Google Scholar] [CrossRef]
- Gaikwad, S.M.; Thakur, B.; Sakpal, A.; Singh, R.K.; Ray, P. Differential Activation of NF-ΚB Signaling Is Associated with Platinum and Taxane Resistance in MyD88 Deficient Epithelial Ovarian Cancer Cells. Int. J. Biochem. Cell Biol. 2015, 61, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.; Ray, P. P53 Loses Grip on PIK3CA Expression Leading to Enhanced Cell Survival during Platinum Resistance. Mol. Oncol. 2016, 10, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Garufi, A.; D’Orazi, V.; Arbiser, J.L.; D’Orazi, G. Gentian Violet Induces Wtp53 Transactivation in Cancer Cells. Int. J. Oncol. 2014, 44, 1084–1090. [Google Scholar] [CrossRef]
- Hanson, G.; Coller, J. Codon Optimality, Bias and Usage in Translation and MRNA Decay. Nat. Rev. Mol. Cell Biol. 2017, 19, 20–30. [Google Scholar] [CrossRef]
- Tuller, T.; Carmi, A.; Vestsigian, K.; Navon, S.; Dorfan, Y.; Zaborske, J.; Pan, T.; Dahan, O.; Furman, I.; Pilpel, Y. An Evolutionarily Conserved Mechanism for Controlling the Efficiency of Protein Translation. Cell 2010, 141, 344–354. [Google Scholar] [CrossRef]
- Galtier, N.; Roux, C.; Rousselle, M.; Romiguier, J.; Figuet, E.; Glémin, S.; Bierne, N.; Duret, L. Codon Usage Bias in Animals: Disentangling the Effects of Natural Selection, Effective Population Size, and GC-Biased Gene Conversion. Mol. Biol. Evol. 2018, 35, 1092–1103. [Google Scholar] [CrossRef]
- Raab, D.; Graf, M.; Notka, F.; Schödl, T.; Wagner, R. The GeneOptimizer Algorithm: Using a Sliding Window Approach to Cope with the Vast Sequence Space in Multiparameter DNA Sequence Optimization. Syst. Synth. Biol. 2010, 4, 215–225. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A General Method Applicable to the Search for Similarities in the Amino Acid Sequence of Two Proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Branchini, B.R.; Ablamsky, D.M.; Murtiashaw, M.H.; Uzasci, L.; Fraga, H.; Southworth, T.L. Thermostable Red and Green Light-Producing Firefly Luciferase Mutants for Bioluminescent Reporter Applications. Anal. Biochem. 2007, 361, 253–262. [Google Scholar] [CrossRef]
- Loening, A.M.; Wu, A.M.; Gambhir, S.S. Red-Shifted Renilla Reniformis Luciferase Variants for Imaging in Living Subjects. Nat. Methods 2007, 4, 641–643. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Valley, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; MacHleidt, T.; et al. Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. ACS Chem. Biol. 2012, 7, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Meier-Soelch, J.; Mayr-Buro, C.; Juli, J.; Leib, L.; Linne, U.; Dreute, J.; Papantonis, A.; Lienhard Schmitz, M.; Kracht, M. Monitoring the Levels of Cellular Nf-Κb Activation States. Cancers 2021, 13, 1–21. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. Regulation of NF-ΚB by TNF Family Cytokines. Semin. Immunol. 2014, 26, 253–266. [Google Scholar] [CrossRef]
- Wang, W.; Mani, A.M.; Wu, Z.-H. DNA Damage-Induced Nuclear Factor-Kappa B Activation and Its Roles in Cancer Progression. J. Cancer Metastasis Treat. 2017, 3, 45. [Google Scholar] [CrossRef] [PubMed]
- Engeland, K. Cell Cycle Regulation: P53-P21-RB Signaling. Cell Death Differ. 2022, 29, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Macleod, K.F.; Sherry, N.; Hannon, G.; Beach, D.; Tokino, T.; Kinzler, K.; Vogelstein, B.; Jacks, T. P53-Dependent and Independent Expression of P21 during Cell Growth, Differentiation, and DNA Damage. Genes Dev. 1995, 9, 935–944. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Guttridge, D.C.; Mayo, M.W.; Albert, S.; Baldwin, J. NF-ΚB Induces Expression of the Bcl-2 Homologue A1/Bfl-1 To Preferentially Suppress Chemotherapy-Induced Apoptosis. Mol. Cell. Biol. 1999, 19, 5923. [Google Scholar] [CrossRef]
- Loening, A.M. Technologies for Imaging with Bioluminescently Labeled Probes, Stanford. 2006. Available online: https://web.stanford.edu/~loening/thesis/Loening_2006_final_normal_quality.pdf (accessed on 24 April 2018).
- Esparza-López, J.; Medina-Franco, H.; Escobar-Arriaga, E.; León-Rodríguez, E.; Zentella-Dehesa, A.; Ibarra-Sánchez, M.J. Doxorubicin Induces Atypical NF-ΚB Activation through c-Abl Kinase Activity in Breast Cancer Cells. J. Cancer Res. Clin. Oncol. 2013, 139, 1625–1635. [Google Scholar] [CrossRef]
- Branchini, B.R.; Southworth, T.L.; Fontaine, D.M.; Kohrt, D.; Florentine, C.M.; Grossel, M.J. A Firefly Luciferase Dual Color Bioluminescence Reporter Assay Using Two Substrates to Simultaneously Monitor Two Gene Expression Events. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef]
- Tamaki, S.; Kitada, N.; Kiyama, M.; Fujii, R.; Hirano, T.; Kim, S.B.; Maki, S. Color-Tunable Bioluminescence Imaging Portfolio for Cell Imaging. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, D.; Ghosh, S. Regulation of the NF-ΚB-Mediated Transcription of Inflammatory Genes. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Widłak, P.; Gramatyka, M.; Kimmel, M. Crosstalk between Stress-Induced NF-ΚB, P53 and HSF1 Signaling Pathways – Review. IFAC Proc. Vol. 2014, 47, 11518–11523. [Google Scholar] [CrossRef]
- Dalmases, A.; González, I.; Menendez, S.; Arpí, O.; Corominas, J.M.; Servitja, S.; Tusquets, I.; Chamizo, C.; Rincón, R.; Espinosa, L.; et al. Deficiency in P53 Is Required for Doxorubicin Induced Transcriptional Activation of NF-ΚB Target Genes in Human Breast Cancer. Oncotarget 2014, 5, 196–210. [Google Scholar] [CrossRef]




| Codon Adaptive Index | GC Content | Codon Frequency Distribution | |
|---|---|---|---|
| Standard | 0.8–1.0 | 30–70% | <30% |
| Luz | 0.71 | 52.15% | 3% |
| hLuz | 0.92 | 56.85% | 0% |
| Nucleotide sequence alignment of Luz and hLuz | ||||
|---|---|---|---|---|
| Max score | Total score | Query cover | E value | Identity |
| 627 | 627 | 96% | 0.0 | 78% |
| Amino acid sequence alignment of Luz and hLuz | ||||
| Max score | Total score | Query cover | E value | Identity |
| 554 | 554 | 100% | 0.0 | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mujawar, A.; Phadte, P.; Palkina, K.A.; Markina, N.M.; Mohammad, A.; Thakur, B.L.; Sarkisyan, K.S.; Balakireva, A.V.; Ray, P.; Yampolsky, I.; et al. Triple Reporter Assay: A Non-Overlapping Luciferase Assay for the Measurement of Complex Macromolecular Regulation in Cancer Cells Using a New Mushroom Luciferase–Luciferin Pair. Sensors 2023, 23, 7313. https://doi.org/10.3390/s23177313
Mujawar A, Phadte P, Palkina KA, Markina NM, Mohammad A, Thakur BL, Sarkisyan KS, Balakireva AV, Ray P, Yampolsky I, et al. Triple Reporter Assay: A Non-Overlapping Luciferase Assay for the Measurement of Complex Macromolecular Regulation in Cancer Cells Using a New Mushroom Luciferase–Luciferin Pair. Sensors. 2023; 23(17):7313. https://doi.org/10.3390/s23177313
Chicago/Turabian StyleMujawar, Aaiyas, Pratham Phadte, Ksenia A. Palkina, Nadezhda M. Markina, Ameena Mohammad, Bhushan L. Thakur, Karen S. Sarkisyan, Anastasia V. Balakireva, Pritha Ray, Ilia Yampolsky, and et al. 2023. "Triple Reporter Assay: A Non-Overlapping Luciferase Assay for the Measurement of Complex Macromolecular Regulation in Cancer Cells Using a New Mushroom Luciferase–Luciferin Pair" Sensors 23, no. 17: 7313. https://doi.org/10.3390/s23177313
APA StyleMujawar, A., Phadte, P., Palkina, K. A., Markina, N. M., Mohammad, A., Thakur, B. L., Sarkisyan, K. S., Balakireva, A. V., Ray, P., Yampolsky, I., & De, A. (2023). Triple Reporter Assay: A Non-Overlapping Luciferase Assay for the Measurement of Complex Macromolecular Regulation in Cancer Cells Using a New Mushroom Luciferase–Luciferin Pair. Sensors, 23(17), 7313. https://doi.org/10.3390/s23177313





