Hybrid Approach to Colony-Forming Unit Counting Problem Using Multi-Loss U-Net Reformulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Multi-Loss U-Net
2.2. Petri Dish Localization
2.3. Hybrid Approach
| Algorithm 1: Proposed hybrid CFU counting approach. | ||||
| Data: input image | ||||
| Result: number N of CFUs | ||||
| 1 | ; | //predict a segmentation output map | ||
| 2 | ; | //find convex hulls of segmented-out regions | ||
| 3 | ; | //fill the inner hollow parts of segmented-out regions | ||
| 4 | ; | //locate a Petri dish, find the extended bezel mask | ||
| 5 | if not empty then | |||
| 6 | ; | //mask everything but inner part of the Petri dish | ||
| 7 | ; | //mask everything but bezel of the Petri dish | ||
| 8 | ||||
| 9 | else | |||
| 10 | ||||
2.4. CFU Counting Application
2.5. Datasets
2.6. Experimental Setup
3. Results
3.1. Main Results
3.2. Stratified Results
3.3. Ablation Study
4. Discussion
- The advantages of the proposed approach are as follows:
- Possibility of seamlessly improving any U-Net architecture;
- Enhanced post-processing techniques that enable improving the contours and regions of segmented-out CFUs;
- Petri dish localization algorithm that enables counting CFUs only on a valid agar plate.
- The disadvantages of the proposed approach are as follows:
- Multiple losses during the training time may require a weighting scheme;
- Increased post-processing time and consumed resources.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brugger, S.D.; Baumberger, C.; Jost, M.; Jenni, W.; Brugger, U.; Mühlemann, K. Automated Counting of Bacterial Colony Forming Units on Agar Plates. PLoS ONE 2012, 7, e33695. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.; Biswas, A.; K, C.; Pal, U. Methods for Rapid Detection of Foodborne Pathogens: An Overview. Am. J. Food Technol. 2011, 6, 87–102. [Google Scholar] [CrossRef]
- Pan, H.; Zhang, Y.; He, G.X.; Katagori, N.; Chen, H. A comparison of conventional methods for the quantification of bacterial cells after exposure to metal oxide nanoparticles. BMC Microbiol 2014, 14, 222. [Google Scholar] [CrossRef] [PubMed]
- ul Maula Khan, A.; Torelli, A.; Wolf, I.; Gretz, N. AutoCellSeg: Robust automatic colony forming unit (CFU)/cell analysis using adaptive image segmentation and easy-to-use post-editing techniques. Sci. Rep. 2018, 8, 7302. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; P.Fischer.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention (MICCAI); LNCS; Springer: Berlin/Heidelberg, Germany, 2015; Volume 9351, pp. 234–241. [Google Scholar]
- Sun, F.; V, A.K.; Yang, G.; Zhang, A.; Zhang, Y. Circle-U-Net: An Efficient Architecture for Semantic Segmentation. Algorithms 2021, 14, 159. [Google Scholar] [CrossRef]
- Isensee, F.; Maier-Hein, K.H. An attempt at beating the 3D U-Net. arXiv 2019, arXiv:1908.02182. [Google Scholar]
- Emek Soylu, B.; Guzel, M.S.; Bostanci, G.E.; Ekinci, F.; Asuroglu, T.; Acici, K. Deep-Learning-Based Approaches for Semantic Segmentation of Natural Scene Images: A Review. Electronics 2023, 12, 2730. [Google Scholar] [CrossRef]
- Zhang, L. Machine learning for enumeration of cell colony forming units. Vis. Comput. Ind. Biomed. Art 2022, 5, 26. [Google Scholar] [CrossRef]
- Chen, X.; Lu, L.; Gao, Y. A new concentric circle detection method based on Hough transform. In Proceedings of the 7th International Conference on Computer Science and Education (ICCSE), Melbourne, Australia, 14–17 July 2012; pp. 753–758. [Google Scholar]
- Hao, G.; Min, L.; Feng, H. Improved Self-Adaptive Edge Detection Method Based on Canny. In Proceedings of the 5th International Conference on Intelligent Human-Machine Systems and Cybernetics, Hangzhou, China, 26–27 August 2013; Volume 2, pp. 527–530. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Advances in Neural Information Processing Systems; Pereira, F., Burges, C., Bottou, L., Weinberger, K., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2012; Volume 25. [Google Scholar]
- Yadav, S.S.; Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data 2019, 6, 113. [Google Scholar] [CrossRef]
- Sutton, R.S.; Barto, A.G. Reinforcement Learning: An Introduction, 2nd ed.; The MIT Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Zhou, S.K.; Le, T.H.N.; Luu, K.; Nguyen, H.V.; Ayache, N. Deep reinforcement learning in medical imaging: A literature review. arXiv 2021, arXiv:2103.05115. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2014; pp. 2672–2680. [Google Scholar]
- Ahmad, W.; Ali, H.; Shah, Z.; Azmat, S. A new generative adversarial network for medical images super resolution. Sci. Rep. 2022, 12, 9533. [Google Scholar] [CrossRef] [PubMed]
- Graczyk, K.M.; Pawłowski, J.; Majchrowska, S.; Golan, T. Self-normalized density map (SNDM) for counting microbiological objects. Sci. Rep. 2022, 12, 10583. [Google Scholar] [CrossRef]
- Li, C.; Li, L.; Jiang, H.; Weng, K.; Geng, Y.; Li, L.; Ke, Z.; Li, Q.; Cheng, M.; Nie, W.; et al. YOLOv6: A Single-Stage Object Detection Framework for Industrial Applications. arXiv 2022, arXiv:2209.02976. [Google Scholar]
- Qin, X.; Zhang, Z.; Huang, C.; Dehghan, M.; Zaiane, O.R.; Jagersand, M. U2-Net: Going deeper with nested U-structure for salient object detection. Pattern Recognit. 2020, 106, 107404. [Google Scholar] [CrossRef]
- Krig, S. Computer Vision Metrics: Survey, Taxonomy, and Analysis; Apress OPEN: New York, NY, USA, 2014. [Google Scholar]
- Das, A.; Medhi, A.; Karsh, R.K.; Laskar, R.H. Image splicing detection using Gaussian or defocus blur. In Proceedings of the International Conference on Communication and Signal Processing (ICCSP), Melmaruvathur, India, 6–8 April 2016; pp. 1237–1241. [Google Scholar]
- Abid, A.; Abdalla, A.; Abid, A.; Khan, D.; Alfozan, A.; Zou, J. Gradio: Hassle-Free Sharing and Testing of ML Models in the Wild. arXiv 2019, arXiv:1906.02569. [Google Scholar]
- Majchrowska, S.; Pawłowski, J.; Guła, G.; Bonus, T.; Hanas, A.; Loch, A.; Pawlak, A.; Roszkowiak, J.; Golan, T.; Drulis-Kawa, Z. AGAR a microbial colony dataset for deep learning detection. arXiv 2021, arXiv:2108.01234. [Google Scholar]
- Mohseni Salehi, S.S.; Erdogmus, D.; Gholipour, A. Tversky Loss Function for Image Segmentation Using 3D fully Convolutional Deep Networks. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2017; pp. 379–387. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems, Software. 2015. Available online: tensorflow.org (accessed on 1 June 2023).
- Müller, D.; Kramer, F. MIScnn: A framework for medical image segmentation with convolutional neural networks and deep learning. BMC Med Imaging 2021, 21, 12. [Google Scholar] [CrossRef]
- Merkel, D. Docker: Lightweight linux containers for consistent development and deployment. Linux J. 2014, 2014, 2. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Proceedings of the 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Granada, Spain, 20 September 2018; Springer: Cham, Switzerland, 2018; Volume 11045, pp. 3–11. [Google Scholar]
- Zhang, Z.; Liu, Q. Road Extraction by Deep Residual U-Net. IEEE Geosci. Remote Sens. Lett. 2017, 15, 749–753. [Google Scholar] [CrossRef]
- Kolařík, M.; Burget, R.; Uher, V.; Říha, K.; Dutta, M.K. Optimized High Resolution 3D Dense-U-Net Network for Brain and Spine Segmentation. Appl. Sci. 2019, 9, 404. [Google Scholar] [CrossRef]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Jumutc, V.; Bļizņuks, D.; Lihachev, A. Multi-Path U-Net Architecture for Cell and Colony-Forming Unit Image Segmentation. Sensors 2022, 22, 990. [Google Scholar] [CrossRef] [PubMed]





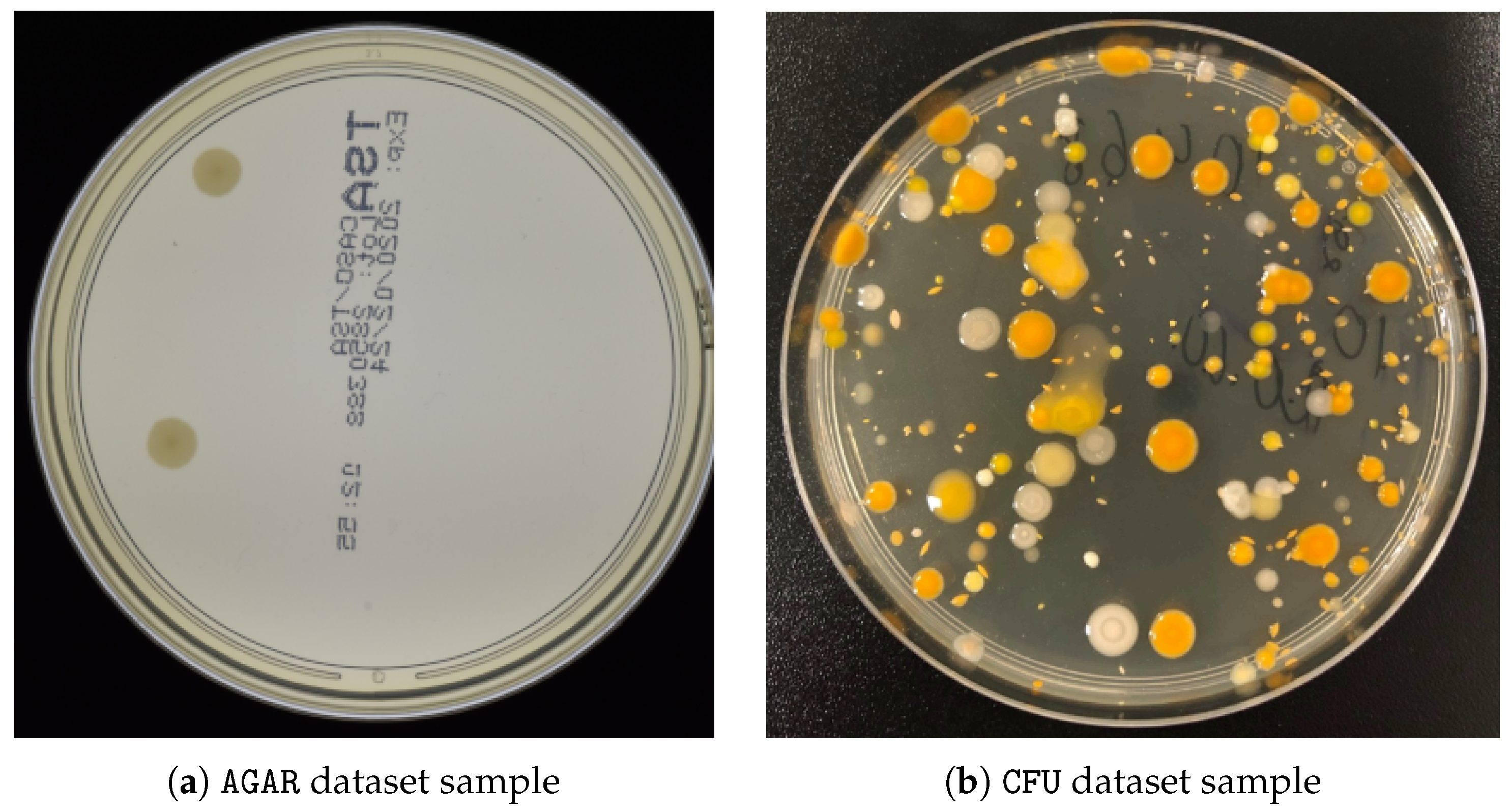

| Dataset | Number of Images | Image Sizes |
|---|---|---|
| AGAR high-resolution | 6990 | 3632 × 4000 × 3 |
| Proprietary CFU | 392 | 3024 × 3024 × 3 |
| Architecture | MAE | sMAPE |
|---|---|---|
| YOLOv6 | 19.69 | 12.41 |
| Single-Loss U-Net++ | 18.54 ± 4.33 | 14.12 ± 3.51 |
| Single-Loss Plain U-Net | 40.44 ± 7.04 | 23.30 ± 3.05 |
| Single-Loss Residual U-Net | 21.24 ± 5.58 | 15.38 ± 5.04 |
| Single-Loss Dense U-Net | 19.14 ± 3.07 | 14.14 ± 2.36 |
| Single-Loss MultiRes U-Net | 15.14 ± 1.70 | 10.37 ± 2.21 |
| Hybrid Single-Loss U-Net++ (ours) | 15.93 ± 2.76 | 11.47 ± 2.41 |
| Hybrid Single-Loss Plain U-Net (ours) | 32.57 ± 3.43 | 19.93 ± 1.63 |
| Hybrid Single-Loss Residual U-Net (ours) | 18.51 ± 3.24 | 13.53 ± 3.88 |
| Hybrid Single-Loss Dense U-Net (ours) | 16.52 ± 2.26 | 12.17 ± 2.14 |
| Hybrid Single-Loss MultiRes U-Net (ours) | 14.13 ± 1.13 | 9.20 ± 1.61 |
| Hybrid Multi-Loss U-Net++ (ours) | 13.06 ± 0.81 | 9.21 ± 0.76 |
| Hybrid Multi-Loss Plain U-Net (ours) | 24.72 ± 2.46 | 16.42 ± 1.49 |
| Hybrid Multi-Loss Residual U-Net (ours) | 12.17 ± 0.22 | 7.73 ± 0.35 |
| Hybrid Multi-Loss Dense U-Net (ours) | 14.10 ± 0.84 | 9.71 ± 0.68 |
| Hybrid Multi-Loss MultiRes U-Net (ours) | 13.42 ± 1.82 | 8.91 ± 1.85 |
| Architecture | MAE | sMAPE |
|---|---|---|
| YOLOv6 | 7.93 | 12.28 |
| Single-Loss U-Net++ | 17.70 ± 5.10 | 36.08 ± 17.39 |
| Single-Loss Plain U-Net | 3.05 ± 0.08 | 3.11 ± 0.22 |
| Single-Loss Residual U-Net | 11.93 ± 1.85 | 17.33 ± 3.83 |
| Single-Loss Dense U-Net | 17.74 ± 6.70 | 27.31 ± 9.84 |
| Single-Loss MultiRes U-Net | 7.55 ± 1.75 | 10.34 ± 2.48 |
| Hybrid Single-Loss U-Net++ (ours) | 17.76 ± 5.07 | 36.20 ± 17.43 |
| Hybrid Single-Loss Plain U-Net (ours) | 3.19 ± 0.07 | 3.37 ± 0.32 |
| Hybrid Single-Loss Residual U-Net (ours) | 11.57 ± 1.96 | 16.98 ± 3.82 |
| Hybrid Single-Loss Dense U-Net (ours) | 17.33 ± 6.44 | 27.48 ± 9.91 |
| Hybrid Single-Loss MultiRes U-Net (ours) | 6.91 ± 1.57 | 9.12 ± 2.18 |
| Hybrid Multi-Loss U-Net++ (ours) | 10.82 ± 1.70 | 18.23 ± 4.30 |
| Hybrid Multi-Loss Plain U-Net (ours) | 1.89 ± 0.04 | 3.25 ± 0.15 |
| Hybrid Multi-Loss Residual U-Net (ours) | 7.67 ± 0.32 | 11.10 ± 1.56 |
| Hybrid Multi-Loss Dense U-Net (ours) | 6.78 ± 0.81 | 10.24 ± 0.57 |
| Hybrid Multi-Loss MultiRes U-Net (ours) | 6.64 ± 1.49 | 8.68 ± 1.78 |
| Architecture | MAE | sMAPE |
|---|---|---|
| Self-Normalized Density Map | 3.65 | 4.05 |
| YOLOv6 | 4.27 | 11.71 |
| Single-Loss U-Net++ | 10.29 ± 3.90 | 34.19 ± 17.42 |
| Single-Loss Plain U-Net | 1.23 ± 0.17 | 2.69 ± 0.23 |
| Single-Loss Residual U-Net | 6.68 ± 1.46 | 15.95 ± 3.78 |
| Single-Loss Dense U-Net | 11.78 ± 6.40 | 25.90 ± 10.12 |
| Single-Loss MultiRes U-Net | 3.84 ± 1.00 | 9.40 ± 2.34 |
| Hybrid Single-Loss U-Net++ (ours) | 10.35 ± 3.87 | 34.32 ± 17.46 |
| Hybrid Single-Loss Plain U-Net (ours) | 1.33 ± 0.17 | 2.95 ± 0.34 |
| Hybrid Single-Loss Residual U-Net (ours) | 6.15 ± 1.47 | 15.48 ± 3.69 |
| Hybrid Single-Loss Dense U-Net (ours) | 11.19 ± 6.15 | 25.97 ± 10.18 |
| Hybrid Single-Loss MultiRes U-Net (ours) | 3.06 ± 0.74 | 8.03 ± 1.98 |
| Hybrid Multi-Loss U-Net++ (ours) | 5.81 ± 1.37 | 16.78 ± 4.29 |
| Hybrid Multi-Loss Plain U-Net (ours) | 0.88 ± 0.05 | 2.17 ± 0.13 |
| Hybrid Multi-Loss Residual U-Net (ours) | 3.61 ± 0.32 | 10.08 ± 1.72 |
| Hybrid Multi-Loss Dense U-Net (ours) | 3.30 ± 0.32 | 9.46 ± 0.44 |
| Hybrid Multi-Loss MultiRes U-Net (ours) | 2.90 ± 0.52 | 7.67 ± 1.48 |
| Architecture | MAE | sMAPE |
|---|---|---|
| YOLOv6 | 10.21 | 14.75 |
| Single-Loss U-Net++ | 15.59 ± 4.94 | 19.11 ± 5.07 |
| Single-Loss Plain U-Net | 38.08 ± 6.22 | 23.30 ± 3.05 |
| Single-Loss Residual U-Net | 18.88 ± 8.46 | 20.79 ± 7.89 |
| Single-Loss Dense U-Net | 17.57 ± 3.81 | 19.28 ± 3.47 |
| Single-Loss MultiRes U-Net | 10.90 ± 2.68 | 13.25 ± 3.47 |
| Hybrid Single-Loss U-Net++ (ours) | 11.44 ± 2.85 | 14.98 ± 3.49 |
| Hybrid Single-Loss Plain U-Net (ours) | 30.84 ± 2.67 | 26.49 ± 2.20 |
| Hybrid Single-Loss Residual U-Net (ours) | 15.33 ± 6.32 | 18.10 ± 6.51 |
| Hybrid Single-Loss Dense U-Net (ours) | 14.30 ± 3.34 | 16.47 ± 3.32 |
| Hybrid Single-Loss MultiRes U-Net (ours) | 9.04 ± 1.73 | 11.39 ± 2.65 |
| Hybrid Multi-Loss U-Net++ (ours) | 8.85 ± 0.90 | 11.87 ± 1.11 |
| Hybrid Multi-Loss Plain U-Net (ours) | 22.41 ± 2.34 | 21.89 ± 2.02 |
| Hybrid Multi-Loss Residual U-Net (ours) | 7.28 ± 0.35 | 9.37 ± 0.65 |
| Hybrid Multi-Loss Dense U-Net (ours) | 10.62 ± 1.15 | 9.71 ± 0.68 |
| Hybrid Multi-Loss MultiRes U-Net (ours) | 9.11 ± 2.85 | 11.24 ± 2.92 |
| Architecture | Multi-Loss only | Multi-Loss (1) | Multi-Loss (2) | |||
|---|---|---|---|---|---|---|
| MAE | sMAPE | MAE | sMAPE | MAE | sMAPE | |
| U-Net++ | 16.00 ± 1.90 | 13.01 ± 2.22 | 13.63 ± 0.87 | 9.93 ± 0.77 | 13.06 ± 0.81 | 9.21 ± 0.76 |
| Plain U-Net | 28.65 ± 2.79 | 18.61 ± 1.58 | 27.84 ± 2.83 | 18.02 ± 1.56 | 24.72 ± 2.46 | 16.42 ± 1.49 |
| Residual U-Net+ | 12.87 ± 0.49 | 8.75 ± 0.59 | 12.75 ± 0.45 | 8.64 ± 0.54 | 12.17 ± 0.22 | 7.73 ± 0.35 |
| Dense U-Net | 15.73 ± 1.17 | 11.08 ± 0.83 | 15.36 ± 1.11 | 10.80 ± 0.79 | 14.10 ± 0.84 | 9.71 ± 0.68 |
| MultiRes U-Net | 14.90 ± 3.08 | 10.33 ± 2.65 | 14.44 ± 2.65 | 9.90 ± 2.25 | 13.42 ± 1.82 | 8.91 ± 1.85 |
| Architecture | Multi-Loss Only | Multi-Loss (1) | Multi-Loss (2) | |||
|---|---|---|---|---|---|---|
| MAE | sMAPE | MAE | sMAPE | MAE | sMAPE | |
| U-Net++ | 10.72 ± 1.70 | 17.97 ± 4.21 | 10.78 ± 1.70 | 18.09 ± 4.23 | 10.82 ± 1.70 | 18.22 ± 4.30 |
| Plain U-Net | 1.74 ± 0.06 | 2.07 ± 0.15 | 1.81 ± 0.05 | 2.20 ± 0.12 | 1.89 ± 0.04 | 2.35 ± 0.15 |
| Residual U-Net+ | 7.76 ± 0.21 | 11.38 ± 1.39 | 7.72 ± 0.24 | 11.23 ± 1.57 | 7.67 ± 0.32 | 11.10 ± 1.56 |
| Dense U-Net | 7.13 ± 1.13 | 10.88 ± 0.91 | 6.92 ± 0.96 | 10.54 ± 0.74 | 6.79 ± 0.81 | 10.24 ± 0.57 |
| MultiRes U-Net | 6.56 ± 1.48 | 8.96 ± 1.69 | 6.60 ± 1.49 | 8.63 ± 1.81 | 6.64 ± 1.49 | 8.68 ± 1.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jumutc, V.; Suponenkovs, A.; Bondarenko, A.; Bļizņuks, D.; Lihachev, A. Hybrid Approach to Colony-Forming Unit Counting Problem Using Multi-Loss U-Net Reformulation. Sensors 2023, 23, 8337. https://doi.org/10.3390/s23198337
Jumutc V, Suponenkovs A, Bondarenko A, Bļizņuks D, Lihachev A. Hybrid Approach to Colony-Forming Unit Counting Problem Using Multi-Loss U-Net Reformulation. Sensors. 2023; 23(19):8337. https://doi.org/10.3390/s23198337
Chicago/Turabian StyleJumutc, Vilen, Artjoms Suponenkovs, Andrey Bondarenko, Dmitrijs Bļizņuks, and Alexey Lihachev. 2023. "Hybrid Approach to Colony-Forming Unit Counting Problem Using Multi-Loss U-Net Reformulation" Sensors 23, no. 19: 8337. https://doi.org/10.3390/s23198337
APA StyleJumutc, V., Suponenkovs, A., Bondarenko, A., Bļizņuks, D., & Lihachev, A. (2023). Hybrid Approach to Colony-Forming Unit Counting Problem Using Multi-Loss U-Net Reformulation. Sensors, 23(19), 8337. https://doi.org/10.3390/s23198337







