Single-Cell Classification Based on Population Nucleus Size Combining Microwave Impedance Spectroscopy and Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Optical Characterization of Cell and Nucleus Size
3.2. Visualization of Electrical Spectra
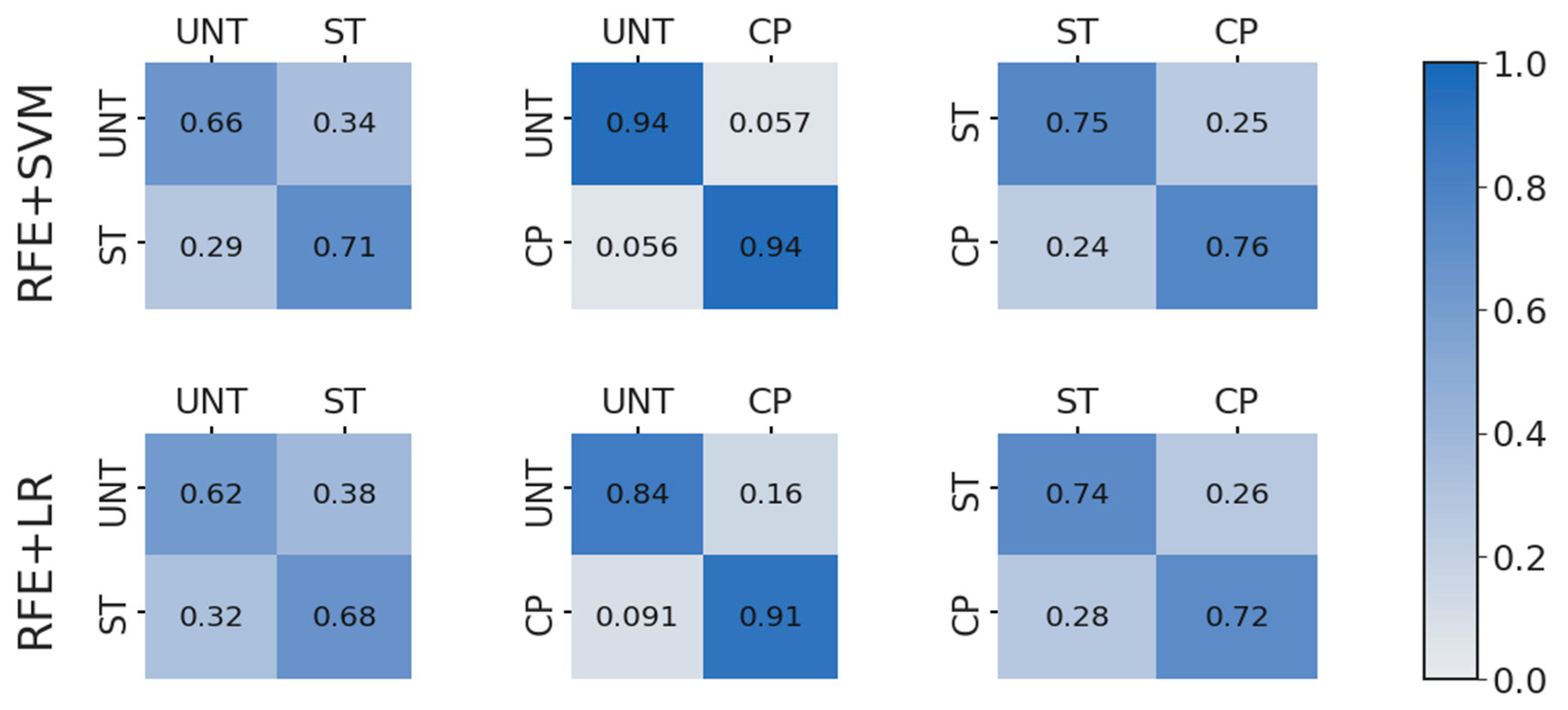
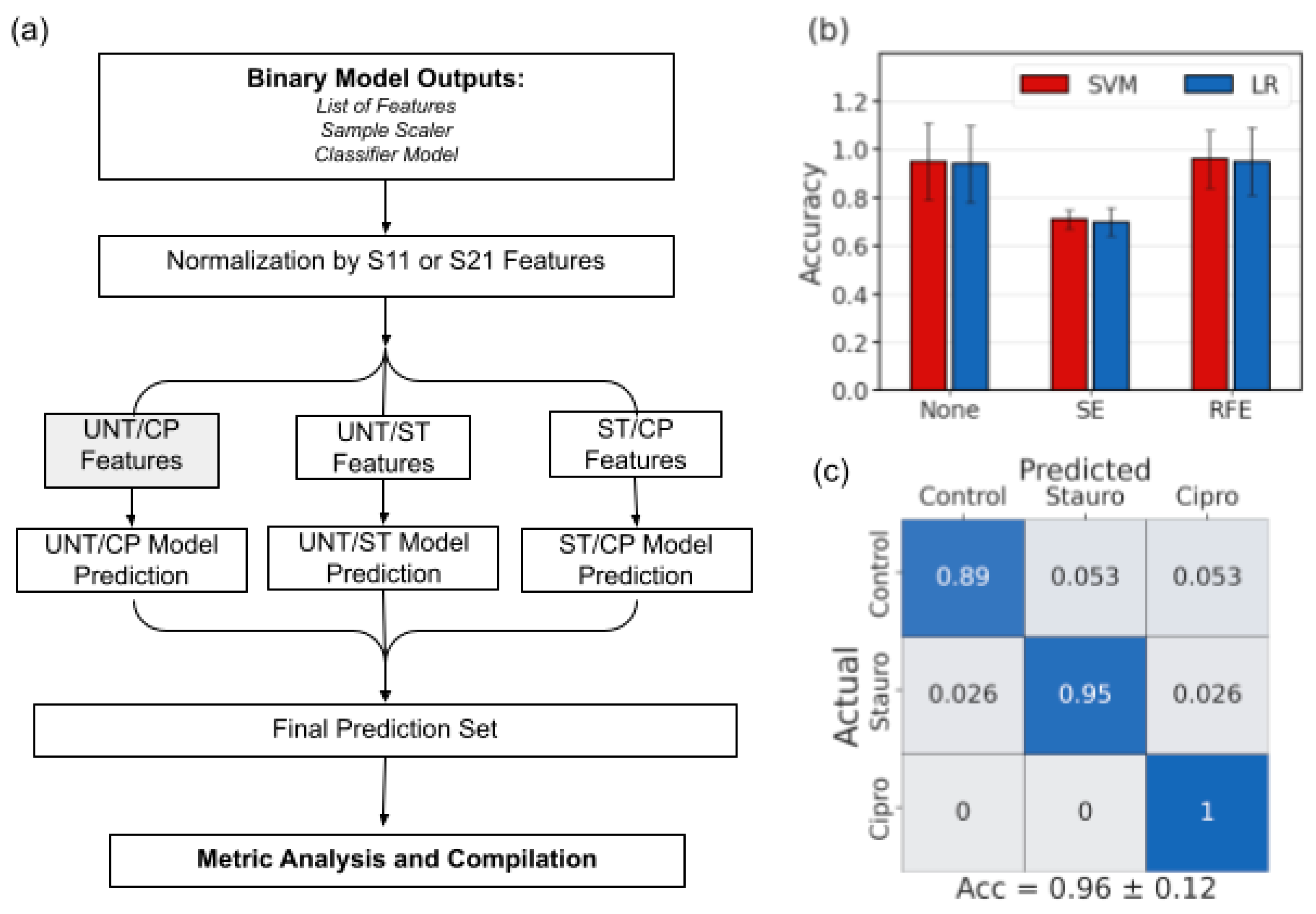
3.3. Binary Feature Selection and Classification
3.4. Multiclass Prediction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A

References
- Sun, Y.; Kwok, Y.C. Polymeric Microfluidic System for DNA Analysis. Anal. Chim. Acta 2006, 556, 80–96. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Barderas, R.; Pedrero, M.; Yáñez-Sedeño, P.; Pingarrón, J.M. Electrochemical Biosensing to Move Forward in Cancer Epigenetics and Metastasis: A Review. Anal. Chim. Acta 2020, 1109, 169–190. [Google Scholar] [CrossRef] [PubMed]
- Matuła, K.; Rivello, F.; Huck, W.T.S. Single-Cell Analysis Using Droplet Microfluidics. Adv. Biosyst. 2020, 4, 1900188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, B.; Xiao, Z. Recent Achievements in Exosomal Biomarkers Detection by Nanomaterials-Based Optical Biosensors—A Review. Anal. Chim. Acta 2020, 1114, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Yang, Y.; Wu, Y.; Liu, S. Review: Multiplexed Profiling of Biomarkers in Extracellular Vesicles for Cancer Diagnosis and Therapy Monitoring. Anal. Chim. Acta 2021, 1175, 338633. [Google Scholar] [CrossRef]
- Deng, Z.; Wang, Y.; Hu, L.; Zhu, Q.; Chen, Y.; Chen, J.J.; Chen, J.; Zhang, T.; Seo, T.S.; Liu, F. A Facile, Rapid, High-Throughput Extracellular Vesicles Analytical Platform for Cancer Detection. Anal. Chim. Acta 2020, 1138, 132–140. [Google Scholar] [CrossRef]
- Yi, C.; Li, C.-W.; Ji, S.; Yang, M. Microfluidics Technology for Manipulation and Analysis of Biological Cells. Anal. Chim. Acta 2006, 560, 1–23. [Google Scholar] [CrossRef]
- Ruedas-Rama, M.J.; Walters, J.D.; Orte, A.; Hall, E.A.H. Fluorescent Nanoparticles for Intracellular Sensing: A Review. Anal. Chim. Acta 2012, 751, 1–23. [Google Scholar] [CrossRef]
- Grenier, K.; Dubuc, D.; Poleni, P.-E.; Kumemura, M.; Toshiyoshi, H.; Fujii, T.; Fujita, H. Integrated Broadband Microwave and Microfluidic Sensor Dedicated to Bioengineering. IEEE Trans. Microw. Theory Techn. 2009, 57, 3246–3253. [Google Scholar] [CrossRef]
- Schwan, H.P. Electrical and Acoustic Properties of Biological Materials and Biomedical Applications. IEEE Trans. Biomed. Eng. 1984, BME-31, 872–878. [Google Scholar] [CrossRef]
- Grenier, K.; Dubuc, D.; Chen, T.; Artis, F.; Chretiennot, T.; Poupot, M.; Fournie, J.-J. Recent Advances in Microwave-Based Dielectric Spectroscopy at the Cellular Level for Cancer Investigations. IEEE Trans. Microw. Theory Techn. 2013, 61, 2023–2030. [Google Scholar] [CrossRef] [Green Version]
- Spencer, D.; Morgan, H. High-Speed Single-Cell Dielectric Spectroscopy. ACS Sens. 2020, 5, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Honrado, C.; Bisegna, P.; Swami, N.S.; Caselli, F. Single-Cell Microfluidic Impedance Cytometry: From Raw Signals to Cell Phenotypes Using Data Analytics. Lab A Chip 2021, 21, 22–54. [Google Scholar] [CrossRef] [PubMed]
- Petchakup, C.; Yang, H.; Gong, L.; He, L.; Tay, H.M.; Dalan, R.; Chung, A.J.; Li, K.H.H.; Hou, H.W. Microfluidic Impedance-Deformability Cytometry for Label-Free Single Neutrophil Mechanophenotyping. Small 2022, 18, 2104822. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, H.; Cheng, X.; Hwang, J.C.M. Sensitivity Analysis for Ultra-Wideband 2-Port Impedance Spectroscopy of a Live Cell. IEEE J. Electromagn. RF Microw. Med. Biol. 2020, 4, 37–44. [Google Scholar] [CrossRef]
- Du, X.; Ladegard, C.; Ma, X.; Cheng, X.; Hwang, J.C.M. Ultra-Wideband Electrical Sensing of Nucleus Size in a Live Cell. In Proceedings of the 2019 49th European Microwave Conference (EuMC), Paris, France, 1–3 October 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 208–211. [Google Scholar]
- Du, X.; Ladegard, C.; Ma, X.; Cheng, X.; Hwang, J.C.M. Broadband Electrical Sensing of Nucleus Size in a Live Cell From 900 Hz to 40 GHz. In Proceedings of the 2020 IEEE MTT-S International Microwave Biomedical Conference (IMBioC), Virtual, 14–17 December 2020; pp. 1–4. [Google Scholar]
- Du, X.; Ferguson, C.; Ma, X.; Cheng, X.; Hwang, J.C.M. Ultra-Wideband Impedance Spectroscopy of the Nucleus in a Live Cell. IEEE J. Electromagn. RF Microw. Med. Biol. 2022, 6, 267–272. [Google Scholar] [CrossRef]
- Zink, D.; Fischer, A.H.; Nickerson, J.A. Nuclear Structure in Cancer Cells. Nat. Rev. Cancer 2004, 4, 677–687. [Google Scholar] [CrossRef]
- Hu, Y.; Hase, T.; Li, H.P.; Prabhakar, S.; Kitano, H.; Ng, S.K.; Ghosh, S.; Wee, L.J.K. A Machine Learning Approach for the Identification of Key Markers Involved in Brain Development from Single-Cell Transcriptomic Data. BMC Genom. 2016, 17, 1025. [Google Scholar] [CrossRef] [Green Version]
- Saadatpour, A.; Lai, S.; Guo, G.; Yuan, G.-C. Single-Cell Analysis in Cancer Genomics. Trends Genet. 2015, 31, 576–586. [Google Scholar] [CrossRef] [Green Version]
- Nassar, M.; Doan, M.; Filby, A.; Wolkenhauer, O.; Fogg, D.K.; Piasecka, J.; Thornton, C.A.; Carpenter, A.E.; Summers, H.D.; Rees, P.; et al. Label-Free Identification of White Blood Cells Using Machine Learning. Cytom. Part A 2019, 95, 836–842. [Google Scholar] [CrossRef]
- Zhang, H.; Shao, X.; Peng, Y.; Teng, Y.; Saravanan, K.M.; Zhang, H.; Li, H.; Wei, Y. A Novel Machine Learning Based Approach for IPS Progenitor Cell Identification. PLoS Comput. Biol. 2019, 15, e1007351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Nowak, C.M.; Pham, U.; Nguyen, K.; Bleris, L. Cell Morphology-Based Machine Learning Models for Human Cell State Classification. Npj Syst. Biol. Appl. 2021, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, K.; Chen, D.; Fan, B.; Xu, Y.; Ye, Y.; Wang, J.; Chen, J.; Huang, C. Development of Microfluidic Impedance Cytometry Enabling the Quantification of Specific Membrane Capacitance and Cytoplasm Conductivity from 100,000 Single Cells. Biosens. Bioelectron. 2018, 111, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Shojaei-Baghini, E.; Wang, C.; Sun, Y. Microfluidic Characterization of Specific Membrane Capacitance and Cytoplasm Conductivity of Singlecells. Biosens. Bioelectron. 2013, 42, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Cheng, Z.; Chai, H.; He, W.; Huang, L.; Wang, W. Neural Network-Enhanced Real-Time Impedance Flow Cytometry for Single-Cell Intrinsic Characterization. Lab A Chip 2022, 22, 240–249. [Google Scholar] [CrossRef]
- Joshi, K.; Javani, A.; Park, J.; Velasco, V.; Xu, B.; Razorenova, O.; Esfandyarpour, R. A Machine Learning-Assisted Nanoparticle-Printed Biochip for Real-Time Single Cancer Cell Analysis. Adv. Biosyst. 2020, 4, 2000160. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, D.; Luo, Y.; Li, H.; Deng, B.; Huang, S.-B.; Chiu, T.-K.; Wu, M.-H.; Long, R.; Hu, H.; et al. A Microfluidic System for Cell Type Classification Based on Cellular Size-Independent Electrical Properties. Lab A Chip 2013, 13, 2272. [Google Scholar] [CrossRef]
- Ahuja, K.; Rather, G.M.; Lin, Z.; Sui, J.; Xie, P.; Le, T.; Bertino, J.R.; Javanmard, M. Toward Point-of-Care Assessment of Patient Response: A Portable Tool for Rapidly Assessing Cancer Drug Efficacy Using Multifrequency Impedance Cytometry and Supervised Machine Learning. Microsyst. Nanoeng. 2019, 5, 34. [Google Scholar] [CrossRef] [Green Version]
- Boateng, E.Y.; Otoo, J.; Abaye, D.A. Basic Tenets of Classification Algorithms K-Nearest-Neighbor, Support Vector Machine, Random Forest and Neural Network: A Review. JDAIP 2020, 08, 341–357. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Cramer, J.S. The Origins of Logistic Regression; Tinbergen Institute Discussion Papers 02-119/4; Tinbergen Institute: Amsterdam, The Netherlands, 2002. [Google Scholar]
- Wang, X.R.; Lizier, J.T.; Berna, A.Z.; Bravo, F.G.; Trowell, S.C. Human Breath-Print Identification by E-Nose, Using Information-Theoretic Feature Selection Prior to Classification. Sensors and Actuators B: Chemical 2015, 217, 165–174. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Yu, H.; Zeng, X.; Zhang, D.; Gong, J.; Tian, L.; Qian, J.; Zhao, L.; Zhang, S.; Liu, R. Lung Cancer Detection via Breath by Electronic Nose Enhanced with a Sparse Group Feature Selection Approach. Sens. Actuators B Chem. 2021, 339, 129896. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. Mach. Learn. Python 6 2011, 12, 2825–2830. [Google Scholar]
- Song, H.; Wang, Y.; Rosano, J.M.; Prabhakarpandian, B.; Garson, C.; Pant, K.; Lai, E. A Microfluidic Impedance Flow Cytometer for Identification of Differentiation State of Stem Cells. Lab A Chip 2013, 13, 2300. [Google Scholar] [CrossRef]
- Guo, J.; Chen, Z.; Ban, Y.-L.; Kang, Y. Precise Enumeration of Circulating Tumor Cells Using Support Vector Machine Algorithm on a Microfluidic Sensor. IEEE Trans. Emerg. Top. Comput. 2017, 5, 518–525. [Google Scholar] [CrossRef]
- Salahi, A.; Honrado, C.; Rane, A.; Caselli, F.; Swami, N.S. Modified Red Blood Cells as Multimodal Standards for Benchmarking Single-Cell Cytometry and Separation Based on Electrical Physiology. Anal. Chem. 2022, 94, 2865–2872. [Google Scholar] [CrossRef]
- Zhou, Y.; Basu, S.; Laue, E.; Seshia, A.A. Single Cell Studies of Mouse Embryonic Stem Cell (MESC) Differentiation by Electrical Impedance Measurements in a Microfluidic Device. Biosens. Bioelectron. 2016, 81, 249–258. [Google Scholar] [CrossRef] [Green Version]
- Honrado, C.; McGrath, J.S.; Reale, R.; Bisegna, P.; Swami, N.S.; Caselli, F. A Neural Network Approach for Real-Time Particle/Cell Characterization in Microfluidic Impedance Cytometry. Anal. Bioanal. Chem. 2020, 412, 3835–3845. [Google Scholar] [CrossRef]
- Ren, X.; Ghassemi, P.; Strobl, J.S.; Agah, M. Biophysical Phenotyping of Cells via Impedance Spectroscopy in Parallel Cyclic Deformability Channels. Biomicrofluidics 2019, 13, 044103. [Google Scholar] [CrossRef]
- Honrado, C.; Salahi, A.; Adair, S.J.; Moore, J.H.; Bauer, T.W.; Swami, N.S. Automated Biophysical Classification of Apoptotic Pancreatic Cancer Cell Subpopulations by Using Machine Learning Approaches with Impedance Cytometry. Lab A Chip 2022, 22, 3708–3720. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferguson, C.A.; Hwang, J.C.M.; Zhang, Y.; Cheng, X. Single-Cell Classification Based on Population Nucleus Size Combining Microwave Impedance Spectroscopy and Machine Learning. Sensors 2023, 23, 1001. https://doi.org/10.3390/s23021001
Ferguson CA, Hwang JCM, Zhang Y, Cheng X. Single-Cell Classification Based on Population Nucleus Size Combining Microwave Impedance Spectroscopy and Machine Learning. Sensors. 2023; 23(2):1001. https://doi.org/10.3390/s23021001
Chicago/Turabian StyleFerguson, Caroline A., James C. M. Hwang, Yu Zhang, and Xuanhong Cheng. 2023. "Single-Cell Classification Based on Population Nucleus Size Combining Microwave Impedance Spectroscopy and Machine Learning" Sensors 23, no. 2: 1001. https://doi.org/10.3390/s23021001
APA StyleFerguson, C. A., Hwang, J. C. M., Zhang, Y., & Cheng, X. (2023). Single-Cell Classification Based on Population Nucleus Size Combining Microwave Impedance Spectroscopy and Machine Learning. Sensors, 23(2), 1001. https://doi.org/10.3390/s23021001






