Convolutional Neural Network Model for Variety Classification and Seed Quality Assessment of Winter Rapeseed
Abstract
:1. Introduction
2. Material and Research Method
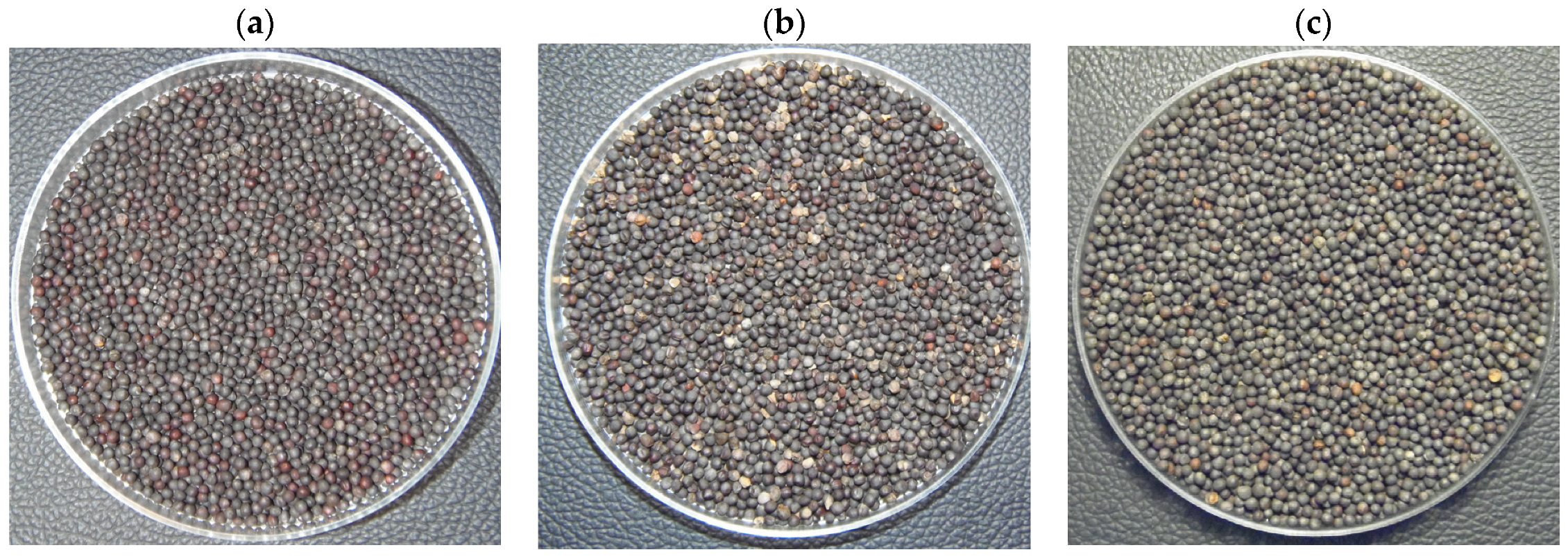
2.1. Data Set Preparation
2.2. Defining Seeds Classification Criteria
2.3. Experimental Set Up
2.4. Loading and Pre-Processing a Data Set
2.5. Multilayer Architecture of CNN Network
3. Results of the Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arrutia, F.; Binnera, E.; Williams, P.; Waldronc, K.W. Oilseeds beyond oil: Press cakes and meals supplying global protein requirements. Trends Food Sci. Technol. 2020, 100, 88–102. [Google Scholar] [CrossRef]
- Campbell, L.; Rempel, C.B.; Wanasundara, J.P. Canola/Rapeseed Protein: Future Opportunities and Directions—Workshop Proceedings of IRC 2015. Plants 2016, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Fairhurst, S.M.; Cole, L.J.; Kocarkova, T.; Jones-Morris, C.; Evans, A.; Jackson, G. Agronomic Traits in Oilseed Rape (Brassica napus) Can Predict Foraging Resources for Insect Pollinators. Agronomy 2021, 11, 440. [Google Scholar] [CrossRef]
- Poisson, E.; Trouverie, J.; Brunel-Muguet, S.; Akmouche, Y.; Pontet, C.; Pinochet, X.; Avice, J.-C. Seed Yield Components and Seed Quality of Oilseed Rape Are Impacted by Sulfur Fertilization and Its Interactions with Nitrogen Fertilization. Front. Plant Sci. 2019, 10, 458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Feng, X.; Wang, J.; Liu, F.; He, Y.; Zhou, W. Mid-infrared spectroscopy combined with chemometrics to detect Sclerotinia stem rot on oilseed rape (Brassica napus L.) leaves. Plant Methods 2017, 13, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Roberts, H.R.; Dodd, I.C.; Hayes, F.; Ashworth, K. Chronic tropospheric ozone exposure reduces seed yield and quality in spring and winter oilseed rape. Agric. For. Meteorol. 2022, 316, 108859. [Google Scholar] [CrossRef]
- USDA, Oilseeds: World Markets and Trade. 2022. Available online: https://www.fas.usda.gov/data/oilseeds-world-markets-and-trade (accessed on 12 May 2022).
- Eurostat. Available online: https://ec.europa.eu/eurostat (accessed on 22 May 2022).
- Namazkar, S.; Stockmarr, A.; Frenck, G.; Egsgaard, H.; Terkelsen, T.; Mikkelsen, T.; Ingvordsen, C.H.; Jørgensen, R.B. Concurrent elevation of CO2, O3 and temperature severely affects oil quality and quantity in rapeseed. J. Exp. Bot. 2016, 67, 4117–4125. [Google Scholar] [CrossRef] [Green Version]
- Niemann, J.; Bocianowski, J.; Wojciechowski, A. Effects of genotype and environment on seed quality traits variability in interspecific cross-derived Brassica lines. Euphytica 2018, 214, 193. [Google Scholar] [CrossRef] [Green Version]
- Niemann, J.; Wojciechowski, A.; Janowicz, J. Broadening the variability of quality traits in rapeseed through interspecific hybridization with an application of immature embryo culture. Biotechnologia 2012, 2, 109–115. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Guan, Z.; Mu, S.; Tang, Q.; Wu, C. Optimization of operating parameter and structure for seed thresher device for rape combine harvester. Nongye Gongcheng Xuebao/Trans. Chin. Soc. Agric. Eng. 2017, 33, 52–57. [Google Scholar] [CrossRef]
- Stępniewski, A.; Szot, B.; Sosnowski, S. Uszkodzenia nasion rzepaku w pozbiorowym procesie obróbki. Acta Agrophysica 2003, 2, 195–203. [Google Scholar]
- Szwed, G. Kształtowanie fizycznych i technologicznych cech nasion rzepaku w modelowych warunkach przechowywania. Acta Agrophysica 2000, 27, 3–116. [Google Scholar]
- Gupta, S.K.; Delseny, M.; Kader, J.C. Advances in botanical research. Incorporating advances in plant pathology. In Rapeseed breeding; Academic Press: Cambridge, MA, USA, 2007; ISBN -13: 978-0-12-374098-4. [Google Scholar]
- Reddy, G.V.P. Integrated Management of Insect Pests on Canola and Other Brassica Oilseed Crops; CPI Group (UK) Ltd.: Croydon, UK, 2017; ISBN -13: 978 1 78064 820 0. [Google Scholar]
- Liu, T.; Tao, B.; Wu, H.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; Fu, T.; Zhu, L.; Shen, J. Bn. YCO affects chloroplast development in Brassica napus L. Crop. J. 2020, 9, 992–1002. [Google Scholar] [CrossRef]
- Kirkegaard, J.A.; Lilley, J.M.; Berry, P.M.; Rondanini, D.P. Chapter 17—Canola. Crop Physiol. Case Hist. Major Crops 2021, 2021, 518–549. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Schmidhuber, J. Deep Learning in Neural Networks: An Overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szegedy, C.; Toshev, A.; Erhan, D. Deep neural networks for object detection. Adv. Neural Inf. Process. Syst. 2013, 2553–2561. [Google Scholar]
- Schroff, F.; Kalenichenko, D.; Philbin, J. Facenet: A unified embedding for face recognition and clustering. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 815–823. [Google Scholar]
- Lemley, J.; Bazrafkan, S.; Corcoran, P. Deep Learning for Consumer Devices and Services: Pushing the limits for machine learning, artificial intelligence, and computer vision. IEEE Consum. Electron. Mag. 2017, 6, 48–56. [Google Scholar] [CrossRef] [Green Version]
- Lemley, J.; Bazrafkan, S.; Corcoran, P. Smart Augmentation Learning an Optimal Data Augmentation Strategy. IEEE Access 2017, 5, 5858–5869. [Google Scholar] [CrossRef]
- Lemley, J.; Bazrafkan, S.; Corcoran, P. Transfer learning of temporal information for driver action classification. In Proceedings of the 28th Modern Artificial Intelligence and Cognitive Science Conference, Fort Wayne, IN, USA, 28–29 April 2017. [Google Scholar] [CrossRef]
- Bordes, A.; Chopra, S.; Weston, J. Question answering with subgraph embeddings. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 26–28 October 2014. [Google Scholar]
- Jean, S.; Cho, K.; Memisevic, R.; Bengio, Y. On using very large target vocabulary for neural machine translation. arXiv 2015, arXiv:1412.2007. [Google Scholar]
- Ma, J.; Sheridan, R.P.; Liaw, A.; Dahl, G.E.; Svetnik, V. Deep Neural Nets as a Method for Quantitative Structure–Activity Relationships. J. Chem. Inf. Model. 2015, 55, 263–274. [Google Scholar] [CrossRef]
- Mikolov, T.; Deoras, A.; Povey, D.; Burget, L.; Cernocky, J. Strategies for training large scale neural network language models. Proc. Autom. Speech Recognit. Underst. 2011, 196–201. [Google Scholar] [CrossRef]
- Sainath, T.N.; Mohamed, A.; Kingsbury, B.; Ramabhadran, B. Deep convolutional neural networks for LVCSR. In Proceedings of the 2013 IEEE International Conference on Acoustics, Speech and Signal Processing, Vancouver, BC, Canada, 26–31 May 2013; pp. 8614–8618. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, X. Machine Translation Quality Assessment of Selected Works of Xiaoping Deng Supported by Digital Humanistic Method. Int. J. Appl. Linguistics Transl. 2021, 7, 59–68. [Google Scholar] [CrossRef]
- Collobert, R.; Weston, J.; Bottou, L.; Karlen, M.; Kavukcuoglu, K.; Kuksa, P. Natural Language Processing (Almost) from Scratch. J. Mach. Learn. Res. 2011, 12, 2493–2537. [Google Scholar]
- van den Oord, A.; Dieleman, S.; Zen, H.; Simonyan, K.; Vinyals, O.; Graves, A.; Kalchbrenner, N.; Senior, A.; Kavukcuoglu, K. WaveNet: A generative model for raw audio. arXiv 2016, arXiv:1609.03499v2. [Google Scholar]
- Zhou, J.; Troyanskaya, O.G. Predicting effects of noncoding variants with deep learning–based sequence model. Nat. Methods 2015, 12, 931–934. [Google Scholar] [CrossRef] [Green Version]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Jermyn, M.; Mok, K.; Mercier, J.; Desroches, J.; Pichette, J.; Saint-Arnaud, K.; Bernstein, L.; Guiot, M.-C.; Petrecca, K.; Leblond, F. Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci. Transl. Med. 2015, 7, 274ra219. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wu, F.; Seo, B.R.; Fischbach, C.; Chen, W.; Hsu, L.; Gourdon, D. Breast cancer cells alter the dynamics of stromal fibronectin-collagen interactions. Matrix Biol. 2016, 60–61, 86–95. [Google Scholar] [CrossRef]
- Levine, S.; Pastor, P.; Krizhevsky, A.; Ibarz, J.; Quillen, D. Learning hand-eye coordination for robotic grasping with deep learning and large-scale data collection. Int. J. Robot. Res. 2017, 37, 421–436. [Google Scholar] [CrossRef]
- Pfeiffer, M.; Schaeuble, M.; Nieto, J.; Siegwart, R.; Cadena, C. From perception to decision: A data-driven approach to end-to-end motion planning for autonomous ground robots. In Proceedings of the 2017 IEEE International Conference on Robotics and Automation (ICRA), Marina Bay Sands, Singapore, 29 May–3 June 2017; pp. 1527–1533. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Davidson, J.; Levine, S.; Sukthankar, R.; Malik, J. Cognitive Mapping and Planning for Visual Navigation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 2616–2625. [Google Scholar]
- Dong, Y.; Tao, J.; Zhang, Y.; Lin, W.; Ai, J. Deep Learning in Aircraft Design, Dynamics, and Control: Review and Prospects. IEEE Trans. Aerosp. Electron. Syst. 2021, 57, 2346–2368. [Google Scholar] [CrossRef]
- Shalev-Shwartz, S.; Shammah, S.; Shashua, A. Safe, Multi-Agent, Reinforcement Learning for Autonomous Driving. arXiv 2016, arXiv:abs/1610.03295. [Google Scholar]
- Bhupendra, M.K.; Miglani, A.; Kankar, P.K. Deep CNN-based damage classification of milled rice grains using a high-magnification image dataset. Comput. Electron. Agric. 2022, 195, 106811. [Google Scholar] [CrossRef]
- Hashim, N.; Onwude, D.I.; Maringgal, B. Chapter 15—Technological advances in postharvest management of food grains. Res. Technol. Adv. Food Sci. 2022, 371–406. [Google Scholar] [CrossRef]
- Ni, J.; Liu, B.; Li, J.; Gao, J.; Yang, H.; Han, Z. Detection of Carrot Quality Using DCGAN and Deep Network with Squeeze-and-Excitation. Food Anal. Methods 2022, 15, 1432–1444. [Google Scholar] [CrossRef]
- Sun, D.; Robbins, K.; Morales, N.; Shu, Q.; Cen, H. Advances in optical phenotyping of cereal crops. Trends Plant Sci. 2021, 27, 191–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qu, M.; Gong, Z.; Cheng, F. Online double-sided identification and eliminating system of unclosed-glumes rice seed based on machine vision. Measurement 2021, 187, 110252. [Google Scholar] [CrossRef]
- Patel, K.K.; Kar, A.; Jha, S.N.; Khan, M.A. Machine vision system: A tool for quality inspection of food and agricultural products. J. Food Sci. Technol. 2012, 49, 123–141. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, S.; Das, A.; Sardana, H.K. Image acquisition techniques for assessment of legume quality. Trends Food Sci. Technol. 2015, 42, 116–133. [Google Scholar] [CrossRef]
- Patrício, D.I.; Rieder, R. Computer vision and artificial intelligence in precision agriculture for grain crops: A systematic review. Comput. Electron. Agric. 2018, 153, 69–81. [Google Scholar] [CrossRef] [Green Version]
- Rybacki, P.; Przygodziński, P.; Osuch, A.; Blecharczyk, A.; Walkowiak, R.; Osuch, E.; Kowalik, I. The Technology of Precise Application of Herbicides in Onion Field Cultivation. Agriculture 2021, 11, 577. [Google Scholar] [CrossRef]
- Rybacki, P.; Przygodziński, P.; Blecharczyk, A.; Kowalik, I.; Osuch, A.; Osuch, E. Strip spraying technology for precise herbicide application in carrot fields. Open Chem. 2022, 20, 287–296. [Google Scholar] [CrossRef]
- Xia, J.; Cao, H.; Yang, Y.; Zhang, W.; Wan, Q.; Xu, L.; Ge, D.; Zhang, W.; Ke, Y.; Huang, B. Detection of waterlogging stress based on hyperspectral images of oilseed rape leaves (Brassica napus L.). Comput. Electron. 2019, 159, 59–68. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, C.; Huang, W.; Liu, F.; He, Y. Application of Hyperspectral Imaging to Detect Sclerotinia sclerotiorum on Oilseed Rape Stems. Sensors 2018, 18, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.R.; Yu, K.Q.; Li, X.; He, Y. Detection of Fungus Infection on Petals of Rapeseed (Brassica napus L.) Using NIR Hyperspectral Imaging. Sci. Rep. 2016, 6, 38878. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, F.; Kong, W.; He, Y. Application of Visible and Near-Infrared Hyperspectral Imaging to Determine Soluble Protein Content in Oilseed Rape Leaves. Sensors 2015, 15, 16576–16588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Liu, F.; Kong, W.; Cui, P.; He, Y.; Zhou, W. Estimation and Visualization of Soluble Sugar Content in Oilseed Rape Leaves Using Hyperspectral Imaging. Trans. ASABE 2016, 59, 1499–1505. [Google Scholar] [CrossRef]
- Bao, Y.; Kong, W.; Liu, F.; Qiu, Z.; He, Y. Detection of Glutamic Acid in Oilseed Rape Leaves Using Near Infrared Spectroscopy and the Least Squares-Support Vector Machine. Int. J. Mol. Sci. 2012, 13, 14106–14114. [Google Scholar] [CrossRef] [Green Version]
- Olivos-Trujillo, M.’; Gajardo, H.A.; Salvo, S.; González, A.; Muñoz, C. Assessing the stability of parameters estimation and prediction accuracy in regression methods for estimating seed oil content in Brassica napus L. using NIR spectroscopy. In Proceedings of the 2015 CHILEAN Conference on Electrical, Electronics Engineering, Information and Communication Technologies (CHILECON), Santiago, Chile, 28–30 October 2015; pp. 25–30. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, F.; He, Y.; Gong, X. Detecting macronutrients content and distribution in oilseed rape leaves based on hyperspectral imaging. Biosyst. Eng. 2013, 115, 56–65. [Google Scholar] [CrossRef]
- Hu, J.; Xu, X.; Liu, L.; Yang, Y. Application of Extreme Learning Machine to Visual Diagnosis of Rapeseed Nutrient Deficiency. In International Forum on Digital TV and Wireless Multimedia Communications; Springer: Singapore, 2018; pp. 238–248. [Google Scholar] [CrossRef]
- Kurtulmuş, F.; Ünal, H. Discriminating rapeseed varieties using computer vision and machine learning. Expert Syst. Appl. 2015, 42, 1880–1891. [Google Scholar] [CrossRef]
- Zou, Q.; Fang, H.; Liu, F.; Kong, W.; He, Y. Comparative Study of Distance Discriminant Analysis and Bp Neural Network for Identification of Rapeseed Cultivars Using Visible/Near Infrared Spectra. In Computer and Computing Technologies in Agriculture IV. CCTA 2010. IFIP Advances in Information and Communication Technology; Li, D., Liu, Y., Chen, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; p. 347. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Guo, X.; Xu, Y.; Zhang, S.; Cheng, X.; Hu, Q.; Wang, W.; Xue, X. Image Recognition of Male Oilseed Rape (Brassica napus) Plants Based on Convolutional Neural Network for UAAS Navigation Applications on Supplementary Pollination and Aerial Spraying. Agriculture 2022, 12, 62. [Google Scholar] [CrossRef]
- Jung, M.; Song, J.S.; Hong, S.; Kim, S.; Go, S.; Lim, Y.P.; Park, J.; Park, S.G.; Kim, Y.-M. Deep Learning Algorithms Correctly Classify Brassica rapa Varieties Using Digital Images. Front. Plant Sci. 2021, 12, 738685. [Google Scholar] [CrossRef] [PubMed]
- Ni, C.; Wang, D.; Vinson, R.; Holmes, M.; Tao, Y. Automatic inspection machine for maize kernels based on deep convolutional neural networks. Biosyst. Eng. 2018, 178, 131–144. [Google Scholar] [CrossRef]
- Lin, P.; Li, X.L.; Chen, Y.M.; He, Y. A deep convolutional neural network architecture for boosting image discrimination accuracy of rice species. Food Bioprocess Technol. 2018, 11, 765–773. [Google Scholar] [CrossRef]
- Zhang, X.; Xun, Y.; Chen, Y. Automated identification of citrus diseases in orchards using deep learning. Biosyst. Eng. 2022, 223, 249–258. [Google Scholar] [CrossRef]
- Bernardes, R.C.; De Medeiros, A.; da Silva, L.; Cantoni, L.; Martins, G.F.; Mastrangelo, T.; Novikov, A.; Mastrangelo, C.B. Deep-Learning Approach for Fusarium Head Blight Detection in Wheat Seeds Using Low-Cost Imaging Technology. Agriculture 2022, 12, 1801. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Hamid, Y.; Wani, S.; Soomro, A.B.; Alwan, A.A.; Gulzar, Y. Smart Seed Classification System based on MobileNetV2 Architecture. In Proceedings of the 2nd International Conference on Computing and Information Technology (ICCIT), Tabuk, Saudi Arabia, 25–27 January 2022; pp. 217–222. [Google Scholar]
- Albarrak, K.; Gulzar, Y.; Hamid, Y.; Mehmood, A.; Soomro, A.B. A Deep Learning-Based Model for Date Fruit Classification. Sustainability 2022, 14, 6339. [Google Scholar] [CrossRef]








| Seed Weight [g] | Research Sample | ||||
|---|---|---|---|---|---|
| Weight Group | Ripe Seeds | Unripe Seeds | Atora | Californium | Graf |
| 1 | 20.000 | 0.000 | atora.0–atora.19 | californium.0–californium.19 | graf.0–graf.19 |
| 2 | 19.839 | 0.161 | atora.20–atora.39 | californium.20–californium.39 | graf.20–graf.39 |
| 3 | 19.677 | 0.323 | atora.40–atora.59 | californium.40–californium.59 | graf.40–graf.59 |
| 4 | 19.516 | 0.484 | atora.60–atora.79 | californium.60–californium.79 | graf.60–graf.79 |
| 5 | 19.355 | 0.645 | atora.80–atora.99 | californium.80–californium.99 | graf.80–graf.99 |
| 6 | 19.194 | 0.807 | atora.100–atora.119 | californium.100–californium.119 | graf.100–graf.119 |
| 7 | 19.032 | 0.968 | atora.120–atora.139 | californium.120–californium.139 | graf.120–graf.139 |
| 8 | 18.871 | 1.129 | atora.140–atora.159 | californium.140–californium.159 | graf.140–graf.159 |
| 9 | 18.710 | 1.290 | atora.160–atora.179 | californium.160–californium.179 | graf.160–graf.179 |
| 10 | 18.548 | 1.452 | atora.180–atora.199 | californium.180–californium.199 | graf.180–graf.199 |
| 11 | 18.387 | 1.613 | atora.200–atora.219 | californium.200–californium.219 | graf.200–graf.219 |
| … | … | … | … | … | … |
| 115 | 1.612 | 18.388 | atora.2280–atora.2299 | californium.2280–californium.2299 | graf.2280–graf.2299 |
| 116 | 1.450 | 18.550 | atora.2300–atora.2319 | californium.2300–californium.2319 | graf.2300–graf.2319 |
| 117 | 1.289 | 18.711 | atora.2320–atora.2339 | californium.2320–californium.2339 | graf.2320–graf.2339 |
| 118 | 1.128 | 18.872 | atora.2340–atora.2359 | californium.2340–californium.2359 | graf.2340–graf.2359 |
| 119 | 0.967 | 19.033 | atora.2360–atora.2379 | californium.2360–californium.2379 | graf.2360–graf.2379 |
| 120 | 0.805 | 19.195 | atora.2380–atora.2399 | californium.2380–californium.2399 | graf.2380–graf.2399 |
| 121 | 0.644 | 19.356 | atora.2400–atora.2419 | californium.2400–californium.2419 | graf.2400–graf.2419 |
| 122 | 0.483 | 19.517 | atora.2420–atora.2439 | californium.2420–californium.2439 | graf.2420–graf.2459 |
| 123 | 0.321 | 19.679 | atora.2440–atora.2459 | californium.2440–californium.2459 | graf.2440–graf.2459 |
| 124 | 0.160 | 19.840 | atora.2460–atora.2479 | californium.2460–californium.2479 | graf.2460–graf.2479 |
| 125 | 0.000 | 20.000 | atora.2480–atora.2499 | californium.2480–californium.2499 | graf.2480–graf.2499 |
| Model CNN | Description |
|---|---|
| RAPESEEDS_AC | Classification of rapeseed varieties Atora F1—Californium |
| RAPESEEDS_CG | Classification of rapeseed varieties Californium—Graf F1 |
| RAPESEEDS_GA | Classification of rapeseed varieties Graf F1—Atora F1 |
| RAPESEEDS_AQ | Evaluation of rape variety Atora F1 |
| RAPESEEDS_CQ | Evaluation of rape variety Californium |
| RAPESEEDS_GQ | Evaluation of rape variety Graf F1 |
| Layer (Type) | Output Shape | Param |
|---|---|---|
| conv2d (Conv2D) | (None, 198, 198, 32) | 896 |
| max_pooling2d (MaxPooling2D) | (None, 99, 99, 32) | 0 |
| dropout (Dropout) | (None, 99, 99, 32) | 0 |
| conv2d_1 (Conv2D) | (None, 97, 97, 64) | 18,496 |
| max_pooling2d_1 (MaxPooling2D) | (None, 48, 48, 64) | 0 |
| dropout_1 (Dropout) | (None, 48, 48, 64) | 0 |
| conv2d_2 (Conv2D) | (None, 46, 46, 128) | 73,856 |
| max_pooling2d_2 (MaxPooling2D) | (None, 23, 23, 128) | 0 |
| dropout_2 (Dropout) | (None, 23, 23, 128) | 0 |
| conv2d_3 (Conv2D) | (None, 21, 21, 128) | 147,584 |
| max_pooling2d_3 (MaxPooling2D) | (None, 10, 10, 128) | 0 |
| dropout_3 (Dropout) | (None, 10, 10, 128) | 0 |
| flatten (Flatten) | (None, 12800) | 0 |
| dense (Dense) | (None, 512) | 6,554,112 |
| dense_1 (Dense) | (None, 1) | 513 |
| Model CNN | Accuracy [%] | Loss | ||
|---|---|---|---|---|
| Training | Validation | Training | Validation | |
| RAPESEEDS_AC | 93.11 | 83.24 | 0.18 | 0.38 |
| RAPESEEDS_CG | 94.19 | 85.60 | 0.17 | 0.43 |
| RAPESEEDS_GA | 90.90 | 83.88 | 0.19 | 0.32 |
| RAPESEEDS_AQ | 93.53 | 80.20 | 0.19 | 0.37 |
| RAPESEEDS_CQ | 90.35 | 80.90 | 0.18 | 0.43 |
| RAPESEEDS_GQ | 92.23 | 81.17 | 0.17 | 0.41 |
| Average | 92.39 | 82.50 | 0.18 | 0.39 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rybacki, P.; Niemann, J.; Bahcevandziev, K.; Durczak, K. Convolutional Neural Network Model for Variety Classification and Seed Quality Assessment of Winter Rapeseed. Sensors 2023, 23, 2486. https://doi.org/10.3390/s23052486
Rybacki P, Niemann J, Bahcevandziev K, Durczak K. Convolutional Neural Network Model for Variety Classification and Seed Quality Assessment of Winter Rapeseed. Sensors. 2023; 23(5):2486. https://doi.org/10.3390/s23052486
Chicago/Turabian StyleRybacki, Piotr, Janetta Niemann, Kiril Bahcevandziev, and Karol Durczak. 2023. "Convolutional Neural Network Model for Variety Classification and Seed Quality Assessment of Winter Rapeseed" Sensors 23, no. 5: 2486. https://doi.org/10.3390/s23052486






