An ECG Stitching Scheme for Driver Arrhythmia Classification Based on Deep Learning
Abstract
1. Introduction
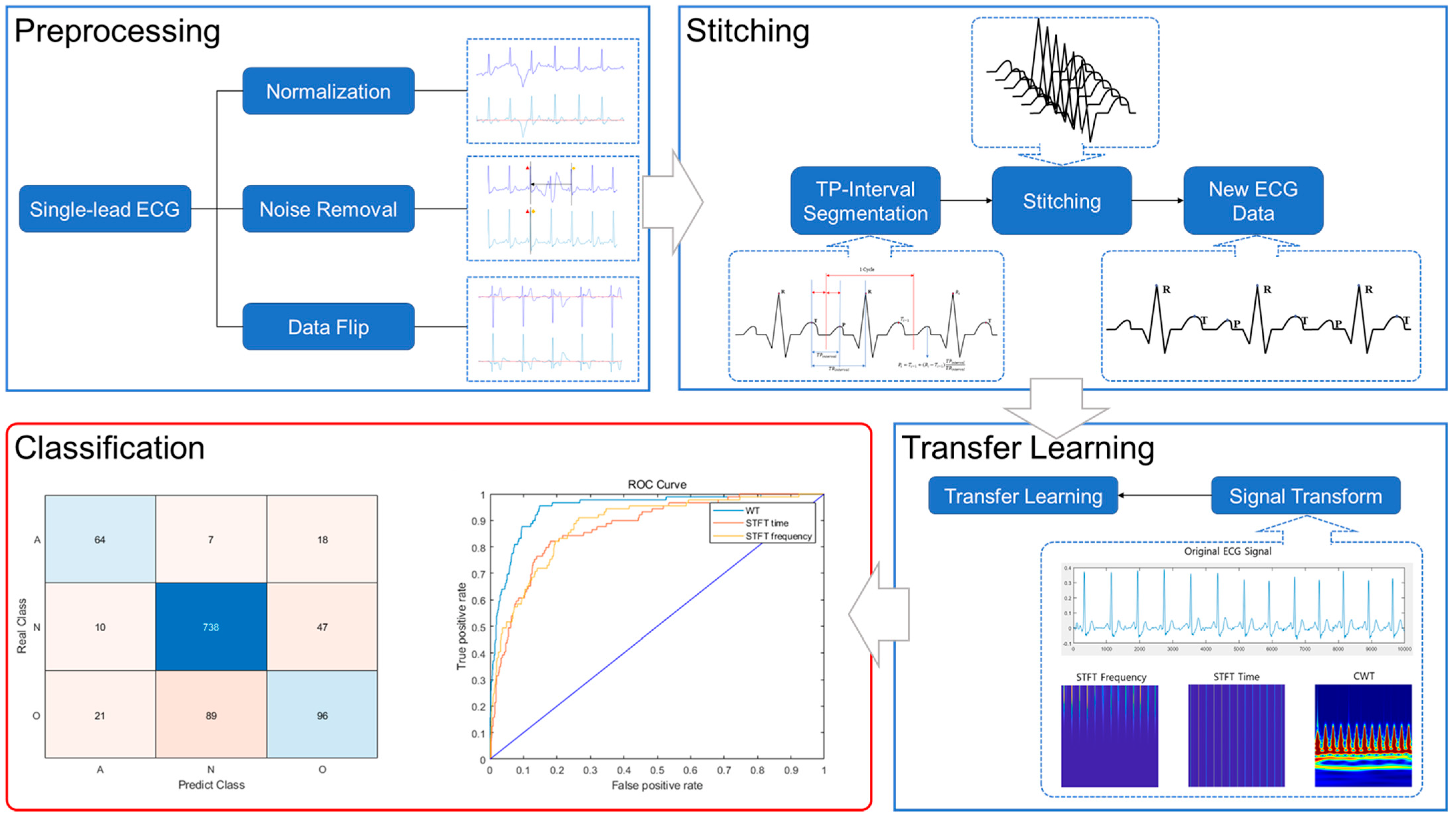
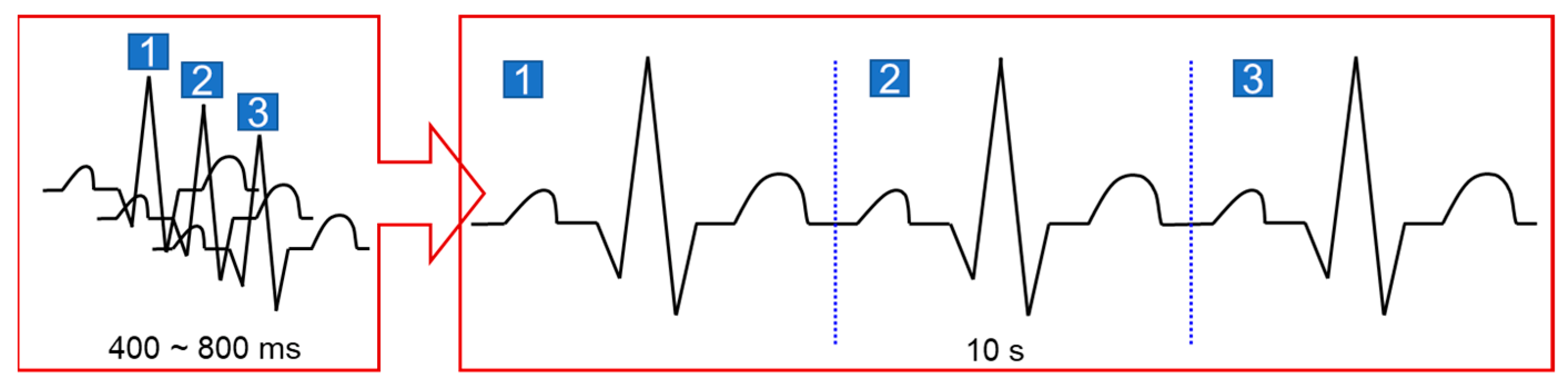
2. ECG Stitching
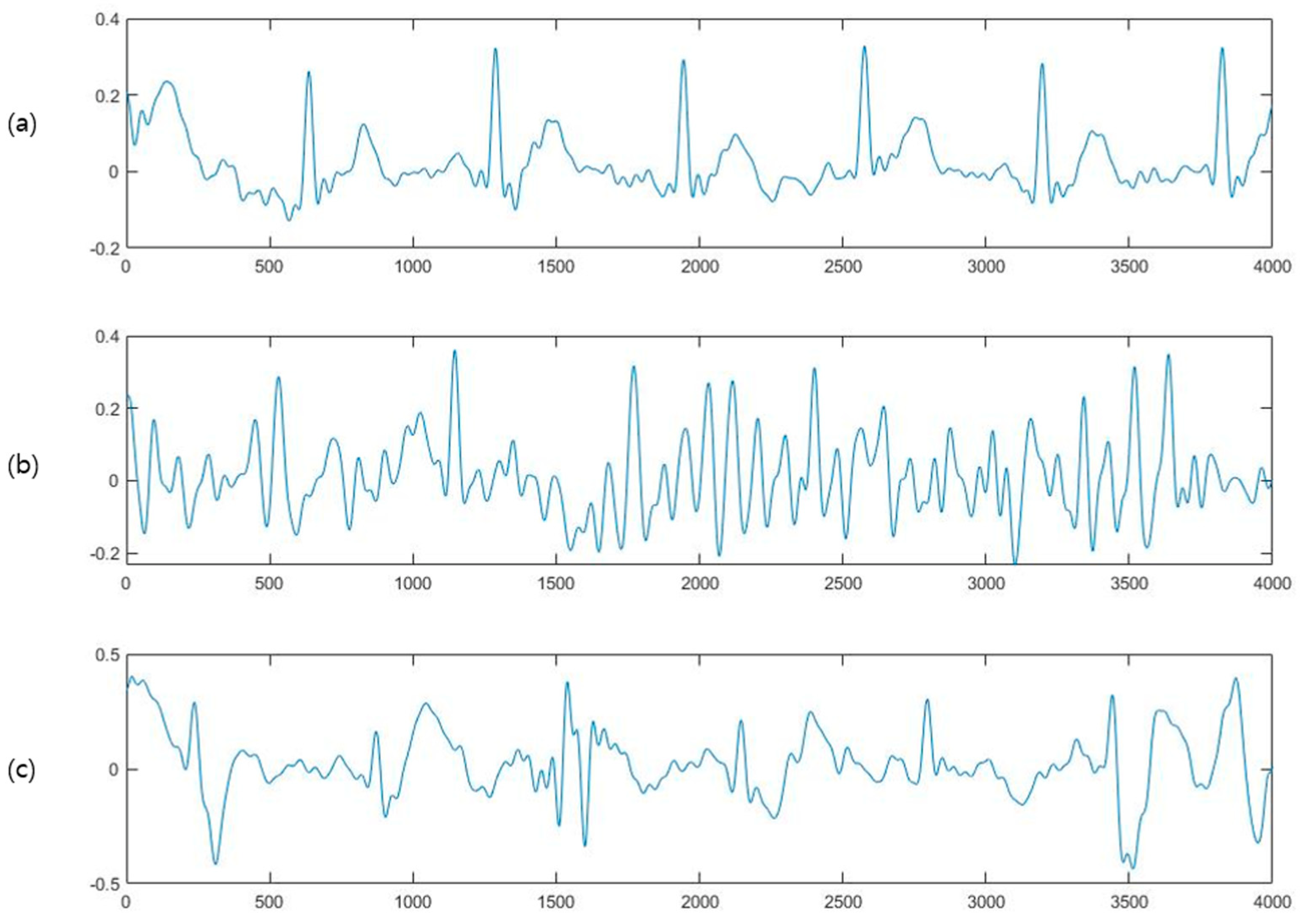
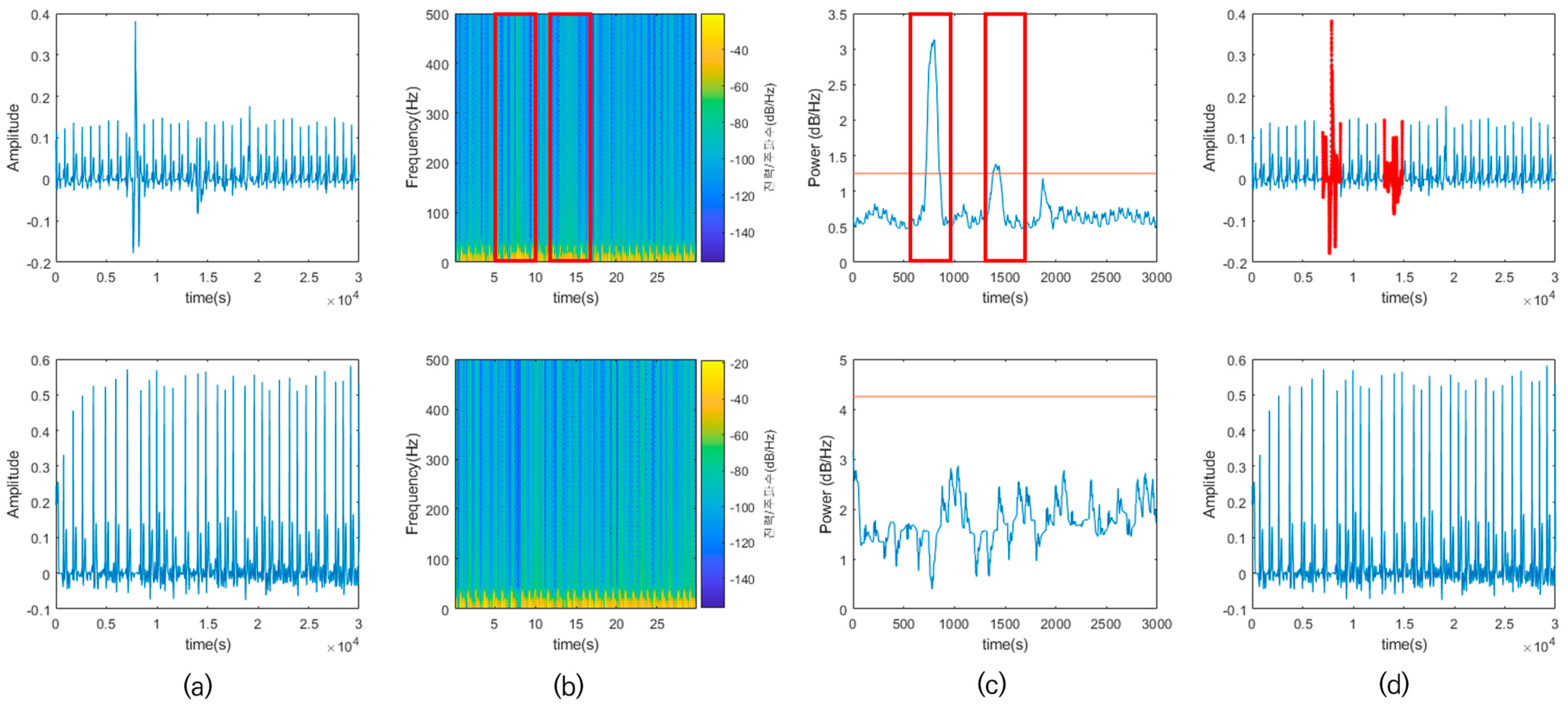
2.1. Preprocessing
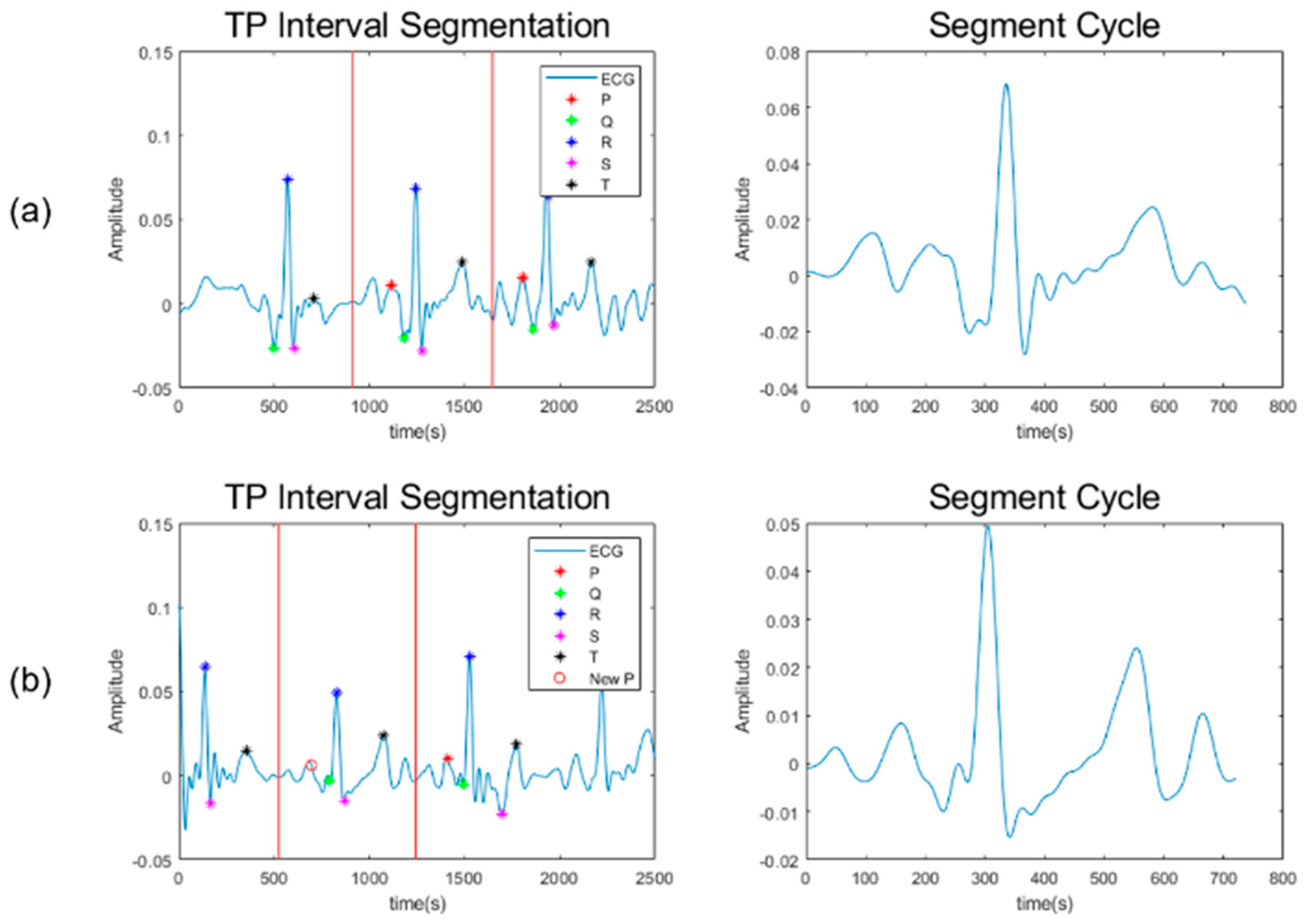
2.2. Signal Stitching
3. Convolutional Neural Network
3.1. Transfer Learning
3.2. Evaluation Matrix
4. Training Setup
4.1. Dataset
4.2. Time–Frequency Analysis
4.3. Training Parameters
4.3.1. Test Dataset
4.3.2. Training Dataset
4.3.3. Initial Training Options
5. Results
5.1. Training Results
5.1.1. Training
5.1.2. Networks
5.1.3. L2 Regularization Rates
5.2. Test Results
- GoogleNet trained using ADAM optimizer.
- CWT-based ECG image set.
- Initial learning rate of 10−4.
- Minibatch size of 64.
- L2 regularization rate of 10−4.
6. Discussion
6.1. Preprocessing Results
6.2. Comparison of the Original and Stitched Data
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Camm, A.J.; Kirchhof, P.; Lip, G.Y.H.; Schotten, U.; Savelieva, I.; Ernst, S.; Gelder, I.C.V.; Al-Attar, N.; Hindricks, G.; Prendergast, B.; et al. Guidelines for the management of atrial fibrillation The Task Force for the Management of Atrial Fibrillation of the European Society of Cardiology (ESC). Eur. Heart J. 2010, 31, 2369–2429. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Clifford, W.; Markham, C.; Deegan, C. Examination of Driver Visual and Cognitive Responses to Billboard Elicited Passive Distraction Using Eye-Fixation Related Potential. Sensors 2021, 21, 1471. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Jia, M.; Hong, Z.; Kwok, A.P.K.; Yan, M. Does augmented-reality head-up display help? A preliminary study on driving performance through a vr-simulated eye movement analysis. IEEE Access 2021, 9, 129951–129964. [Google Scholar] [CrossRef]
- Mao, R.; Li, G.; Hildre, H.; Zhang, H. A survey of eye tracking in automobile and aviation studies: Implications for eye-tracking studies in marine operations. IEEE Trans. Hum. Mach. Syst. 2021, 51, 87–98. [Google Scholar] [CrossRef]
- Lv, C.; Nian, J.; Xu, Y.; Song, B. Compact vehicle driver fatigue recognition technology based on EEG signal. IEEE Trans. Intell. Transp. Syst. 2022, 23, 19753–19759. [Google Scholar] [CrossRef]
- Stancin, I.; Frid, N.; Cifrek, M.; Jovic, A. EEG signal multichannel frequency-domain ratio indices for drowsiness detection based on multicriteria optimization. Sensors 2021, 21, 6932. [Google Scholar] [CrossRef] [PubMed]
- Herry, C.L.; Frasch, M.; Seely, A.J.E.; Wu, H.T. Heart beat classification from single-lead ECG using the synchrosqueezing transform. Physiol. Meas. 2017, 38, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Alfaras, M.; Soriano, M.C.; Ortin, S. A fast machine learning model for ECG-based heartbeat classification and arrhythmia detection. Front. Phys. 2019, 7, 103. [Google Scholar] [CrossRef]
- Hannun, A.Y.; Rajpurkar, P.; Haghpanahi, M.; Tison, G.H.; Bourn, C.; Turakhia, M.P.; Ng, A.Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019, 25, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Parvaneh, S.; Rubin, J.; Babaeizadeh, S.; Xu-Wilson, M. Cardiac arrhythmia detection using deep learning: A review. J. Electrocardiol. 2019, 57, S70–S74. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Lu, Y.; Yang, W.; Wong, S.Y. A study on arrythmia via ECG signal classification using the convolutional neural network. Front. Comput. Neurosci. 2021, 14, 564015. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.; Parvaneh, S.; Rahman, A.; Conroy, B.; Babaeizadeh, S. Densely connected convolutional networks and signal quality analysis to detect atrial fibrillation using short single-lead ECG recordings. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar]
- Mathews, S.M.; Kambhamettu, C.; Barner, K.E. A novel application of deep learning for single-lead ECG classification. Comput. Biol. Med. 2018, 99, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Shang, Y. Modeling physiological data with deep belief networks. Int. J. Inf. Educ. Technol. 2013, 3, 505. [Google Scholar] [PubMed]
- Zhang, W.; Yu, L.; Ye, L.; Zhuang, W.; Ma, F. ECG signal classification with deep learning for heart disease identification. In Proceedings of the 2018 International Conference on Big Data and Artificial Intelligence (BDAI), Beijing, China, 22–24 June 2018; pp. 47–51. [Google Scholar]
- Zubair, M.; Kim, J.; Yoon, C. An automated ECG beat classification system using convolutional neural networks. In Proceedings of the 2016 6th international conference on IT convergence and security (ICITCS), Prague, Czech Republic, 26–29 September 2016; pp. 1–5. [Google Scholar]
- Sereda, I.; Alekseev, S.; Koneva, A.; Kataev, R.; Osipov, G. ECG segmentation by neural networks: Errors and correction. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–7. [Google Scholar]
- Lenis, G.; Pilia, N.; Loewe, A.; Schulze, W.H.; Dössel, O. Comparison of baseline wander removal techniques considering the preservation of ST changes in the ischemic ECG: A simulation study. Comput. Math. Methods Med. 2017, 2017, 9295029. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Johnston, P. A review of electrocardiogram filtering. J. Electrocardiol. 2010, 43, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Choudhury, A.D.; Datta, S.; Puri, C.; Banerjee, R.; Singh, R.; Ukil, A.; Bandyopadhyay, S.; Pal, A.; Khandelwal, S. Detection of atrial fibrillation and other abnormal rhythms from ECG using a multi-layer classifier architecture. Physiol. Meas. 2019, 40, 054006. [Google Scholar] [CrossRef] [PubMed]
- Portet, F. P wave detector with PP rhythm tracking: Evaluation in different arrhythmia contexts. Physiol. Meas. 2008, 29, 141. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, A.A.; Hussain, L.; Awan, I.A.; Abbasi, I.; Majid, A.; Nadeem, M.S.A.; Chaudhary, Q.-A. Detecting prostate cancer using deep learning convolution neural network with transfer learning approach. Cogn. Neurodyn. 2020, 14, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Huang, G.; Liu, Z.; Maaten, L.V.D.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Clifford, G.D.; Liu, C.; Moody, B.; Lehman, L.-W.H.; Silva, I.; Li, Q.; Johnson, A.; Mark, R.G. AF Classification from a Short Single Lead ECG Recording: The PhysioNet/Computing in Cardiology Challenge 2017. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar]
- Wolf, T.; García, C.A.; Castro, D.; Félix, P. Arrhythmia classification from the abductive interpretation of short single-lead ECG records. In Proceedings of the 2017 Computing in Cardiology (Cinc), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar]


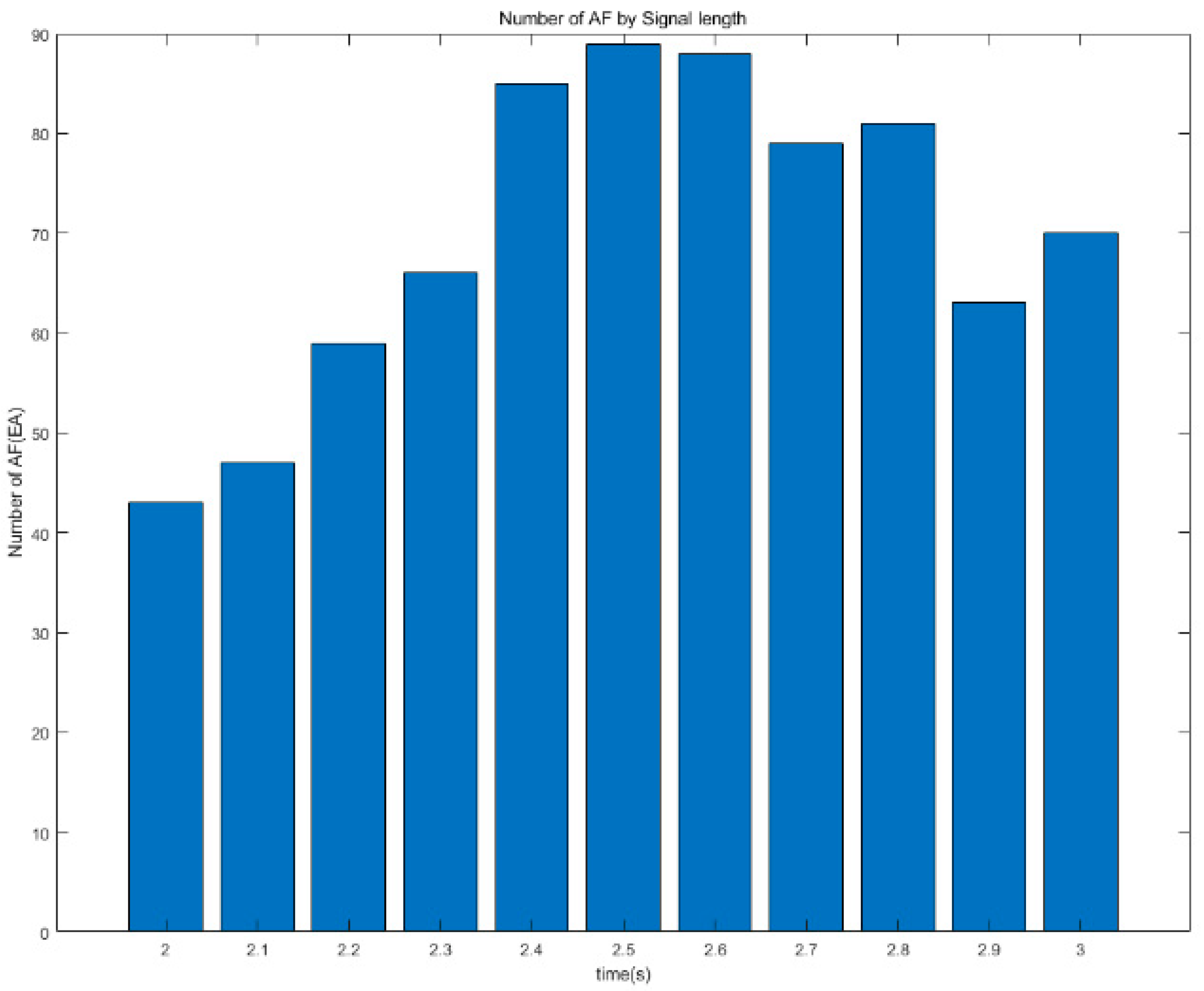


| Time | AF | Normal | Other Rhythms |
|---|---|---|---|
| 2.0 s | 43 | 773 | 228 |
| 2.1 s | 47 | 780 | 221 |
| 2.2 s | 59 | 798 | 203 |
| 2.3 s | 66 | 798 | 203 |
| 2.4 s | 85 | 805 | 196 |
| 2.5 s | 89 | 795 | 206 |
| 2.6 s | 88 | 794 | 207 |
| 2.7 s | 79 | 792 | 209 |
| 2.8 s | 81 | 766 | 235 |
| 2.9 s | 63 | 773 | 228 |
| 3.0 s | 70 | 747 | 254 |
| AF | Normal | Other Rhythms | |
|---|---|---|---|
| AF | TP | FN | FN |
| Normal | FP | TN | FP |
| Other Rhythms | FP | FP | TN |
| Network | Optimizer | Validation Accuracy | Test Accuracy | Validation Loss | |
|---|---|---|---|---|---|
| CWT | GoogleNet | SGDM | 77.46 | 79.36 | 0.5452 |
| ADAM | 81.95 | 82.39 | 0.4757 | ||
| SqueezeNet | SGDM | 76.8 | 78.44 | 0.564 | |
| ADAM | 80.1 | 80.55 | 0.4957 | ||
| ResNet | SGDM | 77.8 | 79.08 | 0.5454 | |
| ADAM | 81.68 | 81.65 | 0.4864 | ||
| DenseNet | SGDM | 78.07 | 78.35 | 0.551 | |
| ADAM | 79.51 | 79.82 | 0.516 | ||
| STFT_F | GoogleNet | SGDM | 72.33 | 75.96 | 0.6836 |
| ADAM | 79.22 | 80.28 | 0.554 | ||
| SqueezeNet | SGDM | 72.67 | 75.87 | 0.6553 | |
| ADAM | 78.9 | 78.81 | 0.5392 | ||
| ResNet | SGDM | 73.89 | 76.88 | 0.7554 | |
| ADAM | 77.31 | 80.37 | 0.5711 | ||
| DenseNet | SGDM | 75.78 | 78.26 | 0.6518 | |
| ADAM | 76.41 | 78.35 | 0.5881 | ||
| STFT_T | GoogleNet | SGDM | 73.94 | 77.89 | 0.6163 |
| ADAM | 79.29 | 80.64 | 0.5331 | ||
| SqueezeNet | SGDM | 75.58 | 78.53 | 0.6078 | |
| ADAM | 78.05 | 80.83 | 0.5382 | ||
| ResNet | SGDM | 75.07 | 77.61 | 0.7499 | |
| ADAM | 76.95 | 80.18 | 0.5805 | ||
| DenseNet | SGDM | 73.38 | 77.61 | 0.7184 | |
| ADAM | 75.63 | 79.54 | 0.5902 |
| Network | Optimizer | Minibatch Size | V Acc | T Acc | V Loss | |
|---|---|---|---|---|---|---|
| CWT | GoogleNet | ADAM | 64 | 81.95 | 82.39 | 0.4757 |
| 128 | 81.47 | 81.83 | 0.486 | |||
| SqueezeNet | ADAM | 64 | 80.1 | 80.55 | 0.4957 | |
| 128 | 79.85 | 79.08 | 0.5118 | |||
| ResNet | ADAM | 64 | 81.68 | 81.65 | 0.4864 | |
| 128 | 80.1 | 77.98 | 0.5197 | |||
| DenseNet | ADAM | 64 | 79.51 | 79.82 | 0.516 | |
| 128 | 80.05 | 78.9 | 0.5437 | |||
| STFT_F | GoogleNet | ADAM | 64 | 79.22 | 80.28 | 0.554 |
| 128 | 78.73 | 80.09 | 0.5573 | |||
| SqueezeNet | ADAM | 64 | 78.9 | 78.81 | 0.5392 | |
| 128 | 78.83 | 79.08 | 0.5324 | |||
| ResNet | ADAM | 64 | 79.18 | 80.37 | 0.5711 | |
| 128 | 79.27 | 79.36 | 0.6009 | |||
| DenseNet | ADAM | 64 | 77.67 | 78.35 | 0.5881 | |
| 128 | 77.62 | 79.17 | 0.5793 | |||
| STFT_T | GoogleNet | ADAM | 64 | 79.29 | 80.64 | 0.5331 |
| 128 | 79.19 | 80.18 | 0.5431 | |||
| SqueezeNet | ADAM | 64 | 78.05 | 80.83 | 0.5382 | |
| 128 | 78.29 | 80.28 | 0.5371 | |||
| ResNet | ADAM | 64 | 79.24 | 80.18 | 0.5805 | |
| 128 | 79.81 | 80.83 | 0.648 | |||
| DenseNet | ADAM | 64 | 78.27 | 79.54 | 0.5902 | |
| 128 | 78.83 | 79.82 | 0.595 |
| Network | Optimizer | Minibatch Size | L2 Rate | V Acc | T Acc | V Loss | |
|---|---|---|---|---|---|---|---|
| CWT | GoogleNet | ADAM | 64 | 10−4 | 81.95 | 82.39 | 0.4757 |
| 10−4 | 78.75 | 79.72 | 0.5101 | ||||
| 0 | 81.00 | 78.44 | 0.4783 | ||||
| SqueezeNet | ADAM | 64 | 10−4 | 80.10 | 80.55 | 0.4957 | |
| 10−2 | 79.95 | 82.52 | 0.5018 | ||||
| 0 | 80.00 | 80.92 | 0.4903 | ||||
| ResNet | ADAM | 64 | 10−4 | 81.68 | 81.65 | 0.4864 | |
| 10−2 | 79.07 | 80.46 | 0.5621 | ||||
| 0 | 82.61 | 81.01 | 0.4790 | ||||
| DenseNet | ADAM | 64 | 10−4 | 79.51 | 79.82 | 0.5160 | |
| 10−2 | 81.47 | 82.02 | 0.4842 | ||||
| 0 | 80.07 | 77.16 | 0.5348 | ||||
| STFT_F | GoogleNet | ADAM | 64 | 10−4 | 79.22 | 80.28 | 0.5540 |
| 10−2 | 77.56 | 79.45 | 0.5549 | ||||
| 0 | 79.10 | 80.00 | 0.5522 | ||||
| SqueezeNet | ADAM | 128 | 10−4 | 78.83 | 79.08 | 0.5324 | |
| 10−2 | 77.14 | 79.19 | 0.5692 | ||||
| 0 | 78.32 | 79.08 | 0.5609 | ||||
| ResNet | ADAM | 64 | 10−4 | 79.18 | 80.37 | 0.5711 | |
| 10−2 | 77.12 | 80.00 | 0.6089 | ||||
| 0 | 79.34 | 81.47 | 0.6017 | ||||
| DenseNet | ADAM | 128 | 10−4 | 76.90 | 79.17 | 0.5793 | |
| 10−2 | 79.17 | 80.00 | 0.5499 | ||||
| 0 | 78.17 | 79.54 | 0.5823 | ||||
| STFT_T | GoogleNet | ADAM | 64 | 10−4 | 79.29 | 80.64 | 0.5331 |
| 10−2 | 76.09 | 79.54 | 0.5814 | ||||
| 0 | 79.02 | 81.19 | 0.5433 | ||||
| SqueezeNet | ADAM | 64 | 10−4 | 78.05 | 80.83 | 0.5382 | |
| 10−2 | 77.00 | 80.09 | 0.5704 | ||||
| 0 | 78.32 | 79.17 | 0.5386 | ||||
| ResNet | ADAM | 128 | 10−4 | 79.81 | 80.83 | 0.6480 | |
| 10−2 | 77.44 | 78.81 | 0.6086 | ||||
| 0 | 78.71 | 80.73 | 0.7514 | ||||
| DenseNet | ADAM | 128 | 10−4 | 78.83 | 79.82 | 0.5950 | |
| 10−2 | 77.81 | 79.91 | 0.6029 | ||||
| 0 | 77.39 | 77.34 | 0.6134 |
| Network | Acc | F1 Score | AUC | |
|---|---|---|---|---|
| CWT | GoogleNet | 82.39 | 0.5950 | 0.9650 |
| SqueezeNet | 80.92 | 0.5066 | 0.9559 | |
| ResNet | 81.01 | 0.5646 | 0.9501 | |
| DenseNet | 82.02 | 0.5673 | 0.9579 | |
| STFT_F | GoogleNet | 80.28 | 0.5141 | 0.9139 |
| SqueezeNet | 79.17 | 0.5171 | 0.9269 | |
| ResNet | 81.47 | 0.5429 | 0.8992 | |
| DenseNet | 80.00 | 0.5018 | 0.9188 | |
| STFT_T | GoogleNet | 80.64 | 0.5288 | 0.9292 |
| SqueezeNet | 79.91 | 0.4996 | 0.9036 | |
| ResNet | 80.83 | 0.5342 | 0.9322 | |
| DenseNet | 79.82 | 0.5153 | 0.9240 |
| Network | Stitched | Original (Non-Stitched) | |||||
|---|---|---|---|---|---|---|---|
| GoogleNet | Predicted Class | Predicted Class | |||||
| A | N | O | A | N | O | ||
| True class | A | 64 | 7 | 18 | 78 | 6 | 5 |
| N | 10 | 738 | 47 | 2 | 778 | 15 | |
| O | 21 | 89 | 96 | 13 | 79 | 114 | |
| Image Set | Acc | F1 | AUC | |
|---|---|---|---|---|
| GoogleNet | Stitched | 82.39 | 0.5950 | 0.9650 |
| Original | 88.99 | 0.7163 | 0.9869 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.H.; Lee, G.; Kim, S.H. An ECG Stitching Scheme for Driver Arrhythmia Classification Based on Deep Learning. Sensors 2023, 23, 3257. https://doi.org/10.3390/s23063257
Kim DH, Lee G, Kim SH. An ECG Stitching Scheme for Driver Arrhythmia Classification Based on Deep Learning. Sensors. 2023; 23(6):3257. https://doi.org/10.3390/s23063257
Chicago/Turabian StyleKim, Do Hoon, Gwangjin Lee, and Seong Han Kim. 2023. "An ECG Stitching Scheme for Driver Arrhythmia Classification Based on Deep Learning" Sensors 23, no. 6: 3257. https://doi.org/10.3390/s23063257
APA StyleKim, D. H., Lee, G., & Kim, S. H. (2023). An ECG Stitching Scheme for Driver Arrhythmia Classification Based on Deep Learning. Sensors, 23(6), 3257. https://doi.org/10.3390/s23063257






