Graph Visualization: Alternative Models Inspired by Bioinformatics
Abstract
:1. Introduction
- Relevance of a visualization model and its focus on one or more application fields, which are characterized by specific data structures and logical relationships inside these structures;
- Heterogeneity of the analyzed models as an opportunity to cover a significant number of visualization models that differ from each other, with aggregation of models based on the principles of their proximity and similarity, including hierarchical, planar, unstructured, temporal, multidimensional, and other models;
- The ability to combine visualization models, including ones with several structures, to more fully display specific data characteristics, including the ability of an analyst to focus simultaneously on several types of information categories, such as overlaying a certain type of diagram on geomaps of the area;
- The presence of practical confirmation of the applicability and usability of models in specific application cases.
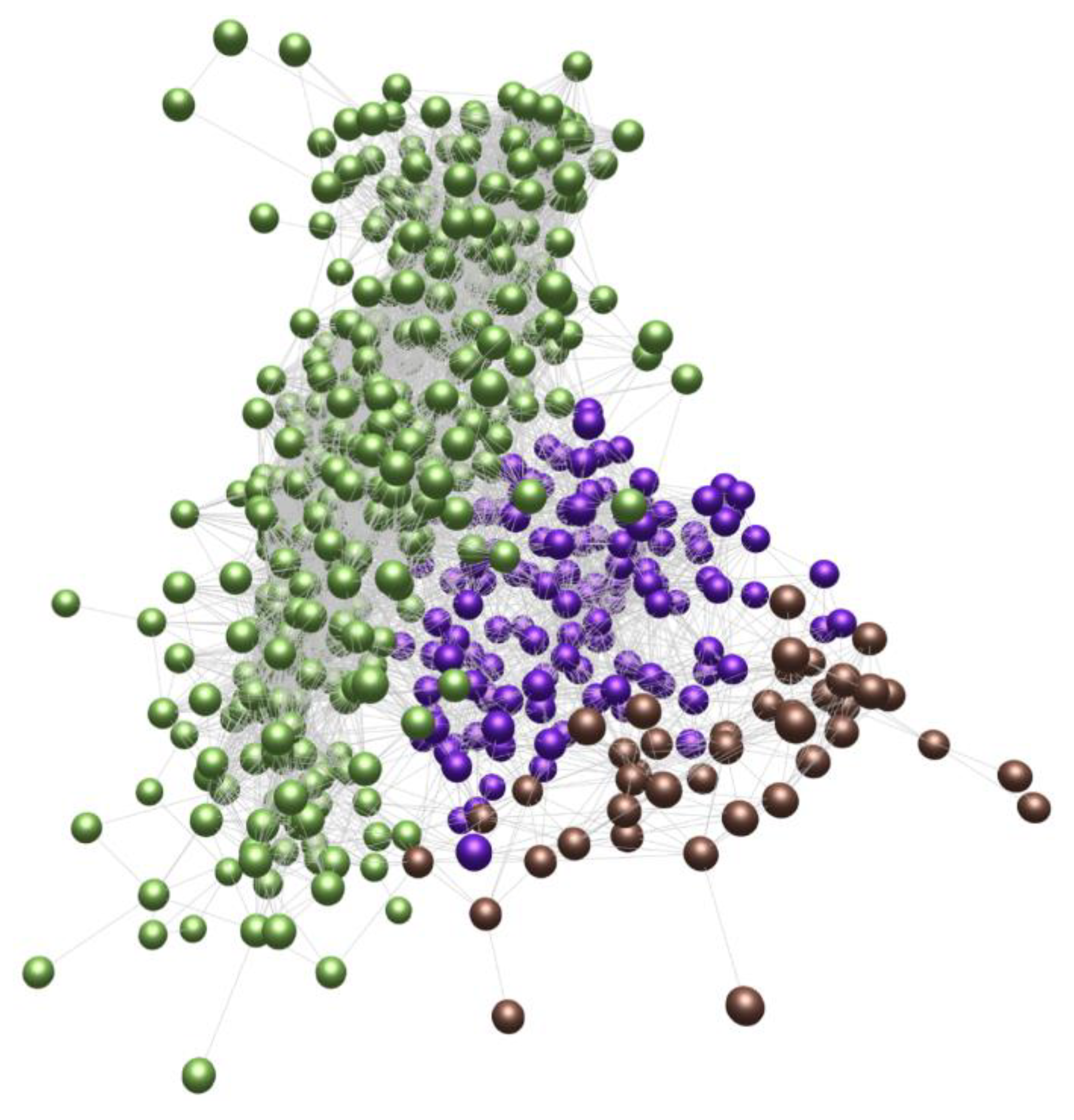
2. State of the Art
3. Alternative Models
3.1. Table Representation
3.2. Tree Maps
3.3. Voronoi Treemaps
3.4. Voronoi Maps
3.5. Chord Diagrams
3.6. Stacked Chord Diagrams
3.7. Voronoi Diagrams
3.8. Trilinear Coordinate Model
3.9. Custom Stacked Models
3.10. Virtual and Augmented Reality
- A group of scientists from South Korea and the United States investigated ways to visualize graph structures in virtual reality on various types of spheres [82].
- Ana Becker (data journalist of The Wall Street Journal) visualized the history of NASDAQ exchange quotes in virtual reality [83], which shows the possibilities of using VR in education and journalism.
- Michal Koutek and Fritz Post developed the MolDRIVE visualization system, which makes it possible to visualize and control experiments in the field of molecular dynamics [84].
- Bob Levy presented the VirtualCove project, which visualizes stock indices in augmented reality [85].
- E-Semble develops emergency simulation programs to train qualified personnel.
- Brown University (Providence, RI, USA) uses virtual reality for various scientific experiments and teaching in psychology, surgery, geology, bioengineering, and other fields [86].
- At the Engenharia Nuclear Institute (Rio de Janeiro, Brazil), the possibilities of using virtual reality to ensure the functioning of nuclear power facilities are being explored [87].
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, X.; Chen, X. Data visualization in smart grid and low-carbon energy systems: A review. Int. Trans. Electr. Energy Syst. 2021, 31, e12889. [Google Scholar] [CrossRef]
- Wiltgen, M.; Holzinger, A. Visualization in Bioinformatics: Protein Structures with Physicochemical and Biological Annotations. In Proceedings of the Central European Multimedia and Virtual Reality Conference, Czech Technical University (CTU), Prague, Czech Republic, 8–10 June 2005; pp. 69–74. [Google Scholar]
- Freemont, P.S. Synthetic biology industry: Data-driven design is creating new opportunities in biotechnology. Emerg. Top. Life Sci. 2019, 3, 651–657. [Google Scholar] [CrossRef]
- O’Donoghue, S.; Gavin, A.-C.; Gehlenborg, N.; Goodsell, D.; Hériché, J.-K.; Nielsen, C.; North, C.; Olson, A.; Procter, J.; Shattuck, D.; et al. Visualizing biological data-now and in the future. Nat. Methods 2010, 7, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Vignesh, U.; Parvathi, R. Biological Big Data Analysis and Visualization: A Survey. Biotechnology: Concepts, Methodologies, Tools, and Applications. IGI Glob. 2020, 653–665. [Google Scholar] [CrossRef]
- Moon, K.; Dijk, D.; Wang, Z.; Burkhardt, D.; Chen, W.; Yim, K.; Elzen, A.; Hirn, M.; Coifman, R.; Ivanova, N.; et al. Visualizing Structure and Transitions for Biological Data Exploration. Available online: https://ssrn.com/abstract=3155891 (accessed on 22 January 2023).
- Raza, K. Application Of Data Mining. Bioinform. Indian J. Comput. Sci. Eng. 2010, 1, 114–118. [Google Scholar]
- Wang Baldonado, M.Q.; Woodruff, A.; Kuchinsky, A. Guidelines for Using Multiple Views in Information Visualization. In Proceedings of the Working Conference on Advanced Visual Interfaces, Palermo, Italy, 23–26 May 2000; pp. 110–119. [Google Scholar] [CrossRef]
- Roberts, J.C. State of the Art: Coordinated Multiple Views in Exploratory Visualization. In Proceedings of the Fifth International Conference on Coordinated and Multiple Views in Exploratory Visualization (CMV 2007), Zurich, Switzerland, 2 July 2007; pp. 61–71. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Luo, Y.; Tang, N. Making data visualization more efficient and effective: A survey. VLDB J. 2020, 29, 93–117. [Google Scholar] [CrossRef]
- Shakeel, H.M.; Iram, S.; Al-Aqrabi, H.; Alsbou’i, T.; Hill, R. A Comprehensive State-of-the-Art Survey on Data Visualization Tools: Research Developments, Challenges and Future Domain Specific Visualization Framework. IEEE Access 2022, 10, 96581–96601. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, Z.; Wang, L.; Sun, G.; Liang, R. Visualization and visual analysis of multimedia data in manufacturing: A survey. Vis. Inform. 2022, 6, 12–21. [Google Scholar] [CrossRef]
- Cakmak, E.; Jäckle, D.; Schreck, T.; Keim, D.; Fuchs, J. Multiscale Visualization: A Structured Literature Analysis. IEEE Trans. Vis. Comput. Graph. 2021, 28, 4918–4929. [Google Scholar] [CrossRef]
- Vaquero, R.; Rzepecki, J.; Friese, K.; Wolter, F.-E. Visualization and User Interaction Methods for Multiscale Biomedical Data. In 3D Multiscale Physiological Human; Springer: London, UK, 2014; pp. 107–133. [Google Scholar] [CrossRef]
- Savinykh, V.; Gospodinov, S.; Kudzh, S.; Tsvetkov, V.; Deshko, I. Semantics of visual models in space research. Russ. Technol. J. 2022, 10, 51–58. [Google Scholar] [CrossRef]
- Neilson, A. Exploring the use of a visual model. In Arts and Mindfulness Education for Human Flourishing; Routledge: Oxfordshire, UK, 2022; pp. 199–212. [Google Scholar] [CrossRef]
- Ke, Z.; Chen, Z.; Wang, H.; Yin, L. A Visual Human-Computer Interaction System Based on Hybrid Visual Model. Secur. Commun. Netw. 2022, 1–13. [Google Scholar] [CrossRef]
- Chiatti, A.; Bardaro, G.; Matteucci, M.; Motta, E. Visual Model Building for Robot Sensemaking: Perspectives, Challenges, and Opportunities. In Proceedings of the Bridge Session on AI and Robotics of the thirty-seventh AAAI conference on Artificial Intelligence (AAAI-23), Washington, WA, USA, 7–14 February 2023. [Google Scholar]
- Hall, D.P.; MacCormick, I.J.C.; Phythian-Adams, A.T.; Rzechorzek, N.M.; Hope-Jones, D.; Cosens, S.; Jackson, S.; Bates, M.G.D.; Collier, D.J.; Hume, D.A.; et al. Network analysis reveals distinct clinical syndromes underlying acute mountain sickness. PLoS ONE 2014, 9, e81229. [Google Scholar] [CrossRef] [Green Version]
- Hooper, S.D.; Bork, P. Medusa: A simple tool for interaction graph analysis. Bioinformatics 2005, 21, 4432–4433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Theocharidis, A.; van Dongen, S.; Enright, A.J.; Freeman, T.C. Network visualization and analysis of gene expression data using BioLayout Express3D. Nat. Protoc. 2009, 4, 1535–1550. [Google Scholar] [CrossRef] [PubMed]
- Breitkreutz, B.J.; Stark, C.; Tyers, M. Osprey: A network visualization system. Genome Biol. 2003, 4, R22. [Google Scholar] [CrossRef] [PubMed]
- Iragne, F.; Nikolski, M.; Mathieu, B.; Auber, D. ProViz: Protein interaction visualization and exploration. Bioinformatics 2004, 21, 272–274. [Google Scholar] [CrossRef] [Green Version]
- Kohler, J.; Baumbach, J.; Taubert, J.; Specht, M.; Skusa, A.; Rüegg, A.; Rawlings, C.; Verrier, P.; Philippi, S.; Notes, A. Graph-based analysis and visualization of experimental results with ONDEX. Bioinformatics 2006, 22, 1383–1390. [Google Scholar] [CrossRef] [Green Version]
- Demir, E.; Babur, O.; Dogrusoz, U.; Gursoy, A.; Nisanci, G.; Cetin-Atalay, R.; Ozturk, M. PATIKA: An integrated visual environment for collaborative construction and analysis of cellular pathways. Bioinformatics 2002, 18, 996–1003. [Google Scholar] [CrossRef] [Green Version]
- Batagelj, V.; Mrvar, A. Pajek-program for large network analysis. Connections 1998, 21, 47–57. [Google Scholar]
- Lopez, D.; Montoya, D.; Ambrose, M.; Lam, L.; Briscoe, L.; Adams, C.; Modlin, R.L.; Pellegrini, M. SaVanT: A web-based tool for the sample-level visualization of molecular signatures in gene expression profiles. BMC Genom. 2017, 18, 824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- iPath3 Pathway Maps Project. Available online: https://pathways.embl.de/ipath3.cgi?map=metabolic (accessed on 1 March 2023).
- Achan, P.; Warrier, A.; Chitturi, B. Biological Data Handling Methods. 2012. Available online: https://pdfs.semanticscholar.org/908e/f3fbd03bc65fd204bb402d827c5088373af1.pdf (accessed on 1 March 2023).
- Beck, F.; Burch, M.; Diehl, S.; Weiskopf, D. A Taxonomy and Survey of Dynamic Graph Visualization. Comput. Graph. Forum 2016, 36, 1–28. [Google Scholar] [CrossRef]
- Majherova, J.; Palasthy, H.; Cernak, I. Visualization of Selected Algorithms of Graph Theory. In Proceedings of the International Conference on E-Learning and E-Technologies in Education, Lodz, Poland, 24–26 September 2012; pp. 17–20. [Google Scholar] [CrossRef]
- Janicke, S.; Heine, C.; Hellmuth, M.; Stadler, P.F.; Scheuermann, G. Visualization of Graph Products. IEEE Trans. Vis. Comput. Graph. 2010, 16, 1082–1089. [Google Scholar] [CrossRef]
- Chevalier, Y.; Fenzl, F.; Kolomeets, M.; Rieke, R.; Chechulin, A.; Kraus, C. Cyberattack detection in vehicles using characteristic functions, artificial neural networks, and visual analysis. Inform. Autom. 2021, 20, 845–868. [Google Scholar] [CrossRef]
- Baehrecke, E.H.; Dang, N.; Babaria, K.; Shneiderman, B. Visualization and analysis of microarray and gene ontology data with treemaps. BMC Bioinform. 2004, 5, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Chandrashekar, P.; Zeng, B.; Sanderford, M.D.; Kumar, S.; Gibson, G. TreeMap: A structured approach to fine mapping of eQTL variants. Bioinformatics 2021, 37, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- López-Ornelas, E.; Abascal-Mena, R. Treemap Visualization: A Hierarchical Method for Discovering User Profiles on Twitter. Comput. Y Sist. 2022, 26, 195–202. [Google Scholar] [CrossRef]
- Aborisade, D.O.; Oyelade, J. HierarchyMap: A Novel Approach to Treemap Visualization of Hierarchical Data. Glob. J. Comput. Sci. Technol. 2010, 5, 77–81. [Google Scholar]
- Jadeja, M.; Shah, K. Tree-map: A visualization tool for large data. In Proceedings of the CEUR Workshop Proceedings, Turin, Italy, 28–29 September 2015; Volume 1393, pp. 9–13. [Google Scholar]
- Shneiderman, B.; Wattenberg, M. Ordered treemap layouts. In Proceedings of the IEEE Symposium on the Information Visualization, Washington, WA, USA, 22–23 October 2001; pp. 73–78. [Google Scholar]
- Bruls, M.; Huizing, K.; van Wijk, J. Squarified treemaps. In Eurographics / IEEE VGTC Symposium on Visualization; The Eurographics Association: Amsterdam, Netherlands, 2000; pp. 33–42. [Google Scholar] [CrossRef]
- Bederson, B.B.; Shneiderman, B.; Wattenberg, M. Ordered and quantum treemaps: Making effective use of 2D space to display hierarchies. AcM Trans. Graph. (TOG) 2002, 21, 833–854. [Google Scholar] [CrossRef] [Green Version]
- Sandy Ressler. Animated Squarified, SliceAndDice and Strip TreeMaps. Available online: https://math.nist.gov/~SRessler/Jit/Examples/Treemap/exampleSAVars2.html (accessed on 1 March 2023).
- Hees, R.V. Stable Voronoi Treemaps for Software System Visualization. Master’s Thesis, Utrecht University, Utrecht, The Netherlands, 2018. [Google Scholar]
- Scheve, M. Impact Landscapes: Supporting the Interpretation and Communication of Life Cycle Assessments with Interactive Voronoi Treemaps. Master’s Thesis, Eindhoven University of Technology, Eindhoven, The Netherlands, 2022. [Google Scholar] [CrossRef]
- Shneiderman, B. A History of Treemap Research at the University of Maryland. Available online: https://www.cs.umd.edu/hcil/treemap-history (accessed on 1 March 2023).
- Kolomeets, M.; Chechulin, A.; Kotenko, I. Visualization Model for Monitoring of Computer Networks Security Based on the Analogue of Voronoi Diagrams. In Proceedings of the International Conference on Availability, Reliability, and Security, Salzburg, Austria, 31 August–2 September 2016; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 141–157. [Google Scholar]
- Blasco-Soplón, L.; Grau, J.; Minguillón, J. Visualization of enrollment data using chord diagrams. In Proceedings of the 10th International Conference on Computer Graphics Theory and Applications (GRAPP 2015), Berlin, Germany, 11–14 March 2015; pp. 511–551. [Google Scholar]
- Caldwell, D.J.; Wu, J.; Casimo, K.; Ojemann, J.G.; Rao, R.P.N. Interactive web application for exploring matrices of neural connectivity. In Proceedings of the 2017 8th International IEEE/EMBS Conference on Neural Engineering (NER), Shanghai, China, 25–28 May 2017; pp. 42–45. [Google Scholar] [CrossRef] [Green Version]
- Powell, S.; Forslund, K.; Szklarczyk, D.; Trachana, K.; Roth, A.; Huerta-Cepas, J.; Gabaldón, T.; Rattei, T.; Creevey, C.; Kuhn, M.; et al. eggNOG v4. 0: Nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014, 42, D231–D239. [Google Scholar] [CrossRef] [Green Version]
- Finnegan, A.; Sao, S.S.; Huchko, M.J. Using a chord diagram to visualize dynamics in contraceptive use: Bringing data into practice. Glob. Health: Sci. Pract. 2019, 7, 598–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poupon, A. Voronoi and Voronoi-related tessellations in studies of protein structure and interaction. Curr. Opin. Struct. Biol. 2004, 14, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; McMains, S. A Voronoi diagram approach for detecting defects in 3D printed fiber-reinforced polymers from microscope images. Comput. Vis. Media 2023, 9, 41–56. [Google Scholar] [CrossRef]
- Figurska, M.; Dawidowicz, A.; Zysk, E. Voronoi Diagrams for Senior-Friendly Cities. Int. J. Environ. Res. Public Health 2022, 19, 7447. [Google Scholar] [CrossRef] [PubMed]
- Adhinugraha, K.; Rahayu, W.; Hara, T.; Taniar, D. Measuring fault tolerance in IoT mesh networks using Voronoi diagram. J. Netw. Comput. Appl. 2022, 199, 103297. [Google Scholar] [CrossRef]
- McConkey, B.J.; Sobolev, V.; Edelman, M. Quantification of protein surfaces, volumes and atom–atom contacts using a constrained Voronoi procedure. Bioinformatics 2002, 18, 1365–1373. [Google Scholar] [CrossRef] [Green Version]
- Gellatly, B.J.; Finney, J.L. Calculation of protein volumes: An alternative to the Voronoi procedure. J. Mol. Biol. 1982, 161, 305–322. [Google Scholar] [CrossRef]
- Goede, A.; Preissner, R.; Frömmel, C. Voronoi cell: New method for allocation of space among atoms: Elimination of avoidable errors in calculation of atomic volume and density. J. Comput. Chem. 1997, 18, 1113–1123. [Google Scholar] [CrossRef]
- Dupuis, F.; Sadoc, J.-F.; Jullien, R.; Angelov, B.; Mornon, J.P. Voro3D: 3D Voronoi tessellations applied to protein structures. Bioinformatics 2005, 21, 1715–1716. [Google Scholar] [CrossRef] [Green Version]
- Boissonnat, J.-D.; Wormser, C.; Yvinec, M. Curved Voronoi Diagrams. In Effective Computational Geometry for Curves and Surfaces; Springer: Berlin/Heidelberg, Germany, 2007; pp. 67–116. [Google Scholar] [CrossRef] [Green Version]
- Lee, D. Two-Dimensional Voronoi Diagrams in the Lp-Metric. J. Assoc. Comput. Mach. 1980, 27, 604–618. [Google Scholar] [CrossRef]
- Whitaker, R.B. Applying Information Visualization to Computer Security Applications; Utah State University: Utah, UT, USA, 2010. [Google Scholar]
- Nunnally, T.; Chi, P.; Abdullah, K.; Uluagac, A.S.; Copeland, J.A.; Beyah, R. P3D: A parallel 3D coordinate visualization for advanced network scans. In Proceedings of the 2013 IEEE International Conference on Communications (ICC), Budapest, Hungary, 9–13 June 2013; pp. 2052–2057. [Google Scholar] [CrossRef]
- Kolomeec, M.; Granadillo, G.G.; Doynikova, E.; Chechulin, A.; Kotenko, I.; Debar, H. Choosing Models for Security Metrics Visualization. In Proceedings of the International Conference on Mathematical Methods, Models, and Architectures for Computer Network, Security, Warsaw, Poland, 28–30 August 2017; Springer: Cham, Germany, 2017; pp. 75–87. [Google Scholar]
- Ying, L.; Shu, X.; Deng, D.; Yang, Y.; Tang, T.; Yu, L.; Wu, Y. MetaGlyph: Automatic generation of metaphoric glyph-based visualization. IEEE Trans. Vis. Comput. Graph. 2022, 29, 331–341. [Google Scholar] [CrossRef]
- Ribarsky, W.; Bolter, J.; Bosch, A.; Teylingen, R. Visualization and Analysis Using Virtual Reality. Comput. Graph. Applications. IEEE 1994, 14, 10–12. [Google Scholar] [CrossRef]
- Ohno, N.; Kageyama, A. Introduction to Virtual Reality Visualization by the CAVE system. Adv. Methods Space Simul. 2007, 167–207. [Google Scholar]
- Maletic, J.; Leigh, J.; Marcus, A. Visualizing Software in an Immersive Virtual Reality Environment. In Proceedings of the ICSE’01 Workshop on Software Visualization, Toronto, Canada, 13–14 May 2001; Society Press: New York, NY, USA, 2001; pp. 12–13. [Google Scholar]
- Lee, S.; El Ali, A.; Wijntjes, M.; Cesar, P. Understanding and Designing Avatar Biosignal Visualizations for Social Virtual Reality Entertainment. In Proceedings of the 2022 CHI Conference on Human Factors in Computing Systems, New Orleans, LA, USA, 30 April–5 May 2022; pp. 1–15. [Google Scholar] [CrossRef]
- Zvarikova, K.; Cug, J.; Hamilton, S. Virtual Human Resource Management in the Metaverse: Immersive Work Environments, Data Visualization Tools and Algorithms, and Behavioral Analytics. Psychosociological Issues Hum. Resour. Manag. 2022, 10, 7–20. [Google Scholar] [CrossRef]
- Huang, C.; Zhang, W.; Xue, L. Virtual reality scene modeling in the context of Internet of Things. Alex. Eng. J. 2022, 61, 5949–5958. [Google Scholar] [CrossRef]
- AlBondakji, L.; Chatzi, A.-M.; Tabar, H.M.; Wesseler, L.-M.; Werner, L. VR-visualization of high-dimensional urban data. In Proceedings of the ECAADE2018, Lodz, Poland, 17–21 September 2018; pp. 773–780. [Google Scholar] [CrossRef]
- Kalkofen, D.; Sandor, C.; White, S.; Schmalstieg, D. Visualization Techniques for Augmented Reality. In Handbook of Augmented Reality; Springer: New York, NY, USA, 2011; pp. 65–98. [Google Scholar] [CrossRef]
- Çöltekin, A.; Griffin, A.L.; Slingsby, A.; Robinson, A.C.; Christophe, S.; Rautenbach, V.; Chen, M.; Pettit, C.; Klippel, A. Geospatial Information Visualization and Extended Reality Displays. In Manual of Digital Earth; Springer: Singapore, 2020; pp. 229–277. [Google Scholar] [CrossRef] [Green Version]
- Tatzgern, M. Situated Visualization in Augmented Reality. Doctoral Thesis to Achieve the University Degree of Doktor der Technischen Wissenschaften. Graz University of Technology. 2015. Available online: https://www.researchgate.net/publication/294088159_Situated_Visualization_in_Augmented_Reality (accessed on 1 March 2023).
- Ramos, F.; Trilles, S.; Torres-Sospedra, J.; Perales, F.J. New Trends in Using Augmented Reality Apps for Smart City Contexts. Int. J. Geo-Inf. 2018, 7, 478. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Nee, A.Y.C.; Ong, S.K. A State-of-the-Art Review of Augmented Reality in Engineering Analysis and Simulation. Multimodal Technol. Interact. 2017, 1, 17. [Google Scholar] [CrossRef]
- Saggio, G.; Ferrari, M. New Trends in Virtual Reality Visualization of 3D Scenarios. In Virtual Reality—Human Computer Interaction, Chapter: 1; InTech Publications: London, UK, 2012; pp. 3–20. [Google Scholar]
- Popovski, F.; Nedelkovski, I.; Mijakovska, S.; Gorica, P. Interactive Scientific Visualization in 3D Virtual Reality Model. Tem J. 2016, 4, 435–440. [Google Scholar] [CrossRef]
- Brennan, M.; Christiansen, L. Virtual Materiality: A Virtual Reality Framework forthe Analysis and Visualization of Cultural Heritage 3D Models. In Proceedings of the 3rd International Congress & Expo Digital Heritage. 2018. Available online: https://www.academia.edu/37852732/Virtual_Materiality_A_Virtual_Reality_Framework_for_the_Analysis_and_Visualization_of_Cultural_Heritage_3D_Models (accessed on 22 January 2023).
- Simpson, M.; Zhao, J.; Klippel, A. Take a Walk: Evaluating Movement Types for Data Visualization in Immersive Virtual Reality. In Proceedings of the Conference: Immersive Analytics: Exploring Future Interaction and Visualization Technologies for Data Analytics (Workshop at IEEE VIS 2017), Phoenix, AZ, USA, 1–6 October 2017. [Google Scholar]
- Kwon, O.H.; Muelder, C.; Lee, K.; Ma, K.L. Spherical layout and rendering methods for immersive graph visualization. In Proceedings of the 2015 IEEE Pacific Visualization Symposium (PacificVis), Hangzhou, China, 14–17 April 2015; pp. 63–67. [Google Scholar]
- Becker, A.A. Designing Virtual Reality Data Visualizations. In Proceedings of the OpenViz Conference, Boston, UK, 25–26 April 2016. [Google Scholar]
- Official Cite of the Project MolDRIVE. Available online: http://graphics.tudelft.nl/~michal/vr_demos (accessed on 22 January 2023).
- Official Cite of the Project VirtualCove. Available online: http://virtualcove.com (accessed on 22 January 2023).
- Dam, A.; Laidlaw, D.H.; Simpson, R.M. Experiments in Immersive Virtual Reality for Scientific Visualization. Comput. Graph. 2002, 4, 535–555. [Google Scholar]
- Da Silva, M.H.; Do Espirito Santo, A.C.; Marins, E.R.; Paula, A.; De Siqueira, L.; Mol, D.M.; Mol, A.C.A. Using virtual reality to support the physical security of nuclear facilities. Prog. Nucl. Energy 2015, 78, 19–24. [Google Scholar] [CrossRef] [Green Version]
- Shan, Q.; Doyle, T.E.; Samavi, R.; Al-Rei, M. Augmented Reality Based Brain Tumor 3D Visualization. Procedia Comput. Sci. 2017, 113, 400–407. [Google Scholar] [CrossRef]
- Alvarado, Y.; Moyano, N.; Quiroga, D.; Fernandez, J.; Guerrero, R. A Virtual Reality Computing Platform for Real Time 3D Visualization. In Proceedings of the XVIII Congreso Argentino de Ciencias de la Computación, Bahia Blanca, Argentina, 12 October 2012. [Google Scholar]
- Kim, Y.S.; Walls, L.A.; Krafft, P.; Hullman, J. A Bayesian cognition approach to improve data visualization. In Proceedings of the 2019 chi conference on human factors in computing systems, Glasgow, UK, 4–9 May 2019; pp. 1–14. [Google Scholar] [CrossRef] [Green Version]
- Vervier, K.; Mahé, P.; Vert, J.-P. MetaVW: Large-Scale Machine Learning for Metagenomics Sequence Classification. Data Min. Syst. Biol. Methods Protoc. Methods Mol. Biol. 2018, 1807, 9–20. [Google Scholar] [CrossRef]
- Macklin, P. Key challenges facing data-driven multicellular systems biology. GigaScience 2019, 8, 1–8. [Google Scholar] [CrossRef]
- Kerren, A.; Kucher, K.; Li, Y.-F.; Schreiber, F. BioVis Explorer: A visual guide for biological data visualization techniques. PLoS ONE 2017, 12, e0187341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frank, J. Visualization of Molecular Machines by Cryo-Electron Microscopy. Vis. Mol. Mach. By Cryo-Electron Microsc. 2011, 20–37. [Google Scholar] [CrossRef]
- Novikova, E.; Bestuzhev, M.; Kotenko, I. Anomaly Detection in the HVAC System Operation by a RadViz Based Visualization-Driven Approach. In Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2020; Volume 11980, pp. 402–418. [Google Scholar] [CrossRef]
- Kolomeets, M.; Chechulin, A.; Zhernova, K.; Kotenko, I.; Gaifulina, D. Augmented reality for visualizing security data for cybernetic and cyberphysical systems. In Proceedings of the 2020 28th Euromicro International Conference on Parallel, Distributed and Network-Based Processing (PDP), Vasteras, Sweden, 11–13 March 2020; pp. 421–428. [Google Scholar]
- Wang, J.; Hazarika, S.; Li, C.; Shen, H.-W. Visualization and Visual Analysis of Ensemble Data: A Survey. IEEE Trans. Vis. Comput. Graph. 2018, 25, 1–20. [Google Scholar] [CrossRef]
- Kolomeec, M.; Chechulin, A.; Kotenko, I. Visual analysis of CAN bus traffic injection using radial bar charts. In Proceedings of the 2018 IEEE Industrial Cyber-Physical Systems, ICPS 2018, The 1st IEEE International Conference on Industrial Cyber-Physical Systems (ICPS2018), Saint Petersburg, Russia, 15–18 May 2018; pp. 841–846. [Google Scholar] [CrossRef]
- Meleshko, A.; Shulepov, A.; Desnitsky, V.; Novikova, E.; Kotenko, I. Visualization Assisted Approach to Anomaly and Attack Detection in Water Treatment Systems. Water 2022, 14, 2342. [Google Scholar] [CrossRef]
- Novikova, E.; Kotenko, I.; Murenin, I. The visual analytics approach for analyzing trajectories of critical infrastructure employers. Energies 2020, 13, 3936. [Google Scholar] [CrossRef]
- Novikova, E.; Kotenko, I. Visualization-Driven Approach to Fraud Detection in the Mobile Money Transfer Services. In Algorithms, Methods, and Applications in Mobile Computing and Communications; IGI Global: Hershey, PA, USA, 2019; pp. 205–236. [Google Scholar] [CrossRef]
- Pronoza, A.; Vitkova, L.; Chechulin, A.; Kotenko, I. Visual analysis of information dissemination channels in social network for protection against inappropriate content. In Proceedings of the Advances in Intelligent Systems and Computing, Proceedings of the 3rd International Scientific Conference “Intelligent Information Technologies for Industry” (IITI’18), Sochi, Russia, 17–21 September 2018; Volume 875, pp. 95–105. [Google Scholar] [CrossRef]
- Kolomeets, M.; Chechulin, A.; Kotenko, I.; Chevalier, Y. A visual analytics approach for the cyber forensics based on different views of the network traffic. J. Wirel. Mob. Netw. Ubiquitous Comput. Dependable Appl. 2018, 9, 57–73. [Google Scholar]
- Demiralp, Ç.; Scheiddegger, C.; Kindlmann, G.; Laidlaw, D.; Heer, J. Visual Embedding: A Model for Visualization. IEEE Comput. Graph. Appl. 2014, 34, 10–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forbes, F. Modelling structured data with probabilistic graphical models. EAS Publ. Ser. 2016, 77, 195–219. [Google Scholar] [CrossRef] [Green Version]
- Jordan, M.; Sejnowski, T. Graphical Models: Foundations of Neural Computation. Pattern Anal. Appl. 2002, 5, 401–402. [Google Scholar] [CrossRef]
- Cevallos, Y.; Oquendo, L.T.; Inca, D.; Arias, C.P.; Renteria, L. Genetic Expression in Biological Systems: A Digital Communication Perspective. Open Bioinform. J. 2019, 12, 45–49. [Google Scholar] [CrossRef] [Green Version]
- Bharne, D.; Kant, P.; Vindal, V. maGUI: A Graphical User Interface for Analysis and Annotation of DNA Microarray Data. Open Bioinform. J. 2019, 12, 40–44. [Google Scholar] [CrossRef]





















| No. | Topology type | Description |
|---|---|---|
| 1 | Unrelated data | A data set that does not present a network (a set of features with no links between them) |
| 2 | Hierarchical networks | Planar networks that can be represented as a tree |
| 3 | Planar networks | Networks for which planarity can be proved and for which a planar image in the plane can be found in polynomial time |
| 4 | Unstructured networks | Networks for which it is impossible to single out a single-valued structure or whose structure is impossible to determine due to the large amount of data that cannot be analyzed in polynomial time |
| 5 | Combined networks | Networks that have several topology levels (a set of objects with links of different types), while different topologies do not have to be of the same type |
| No. | Topology Type | Visualization Model |
|---|---|---|
| 1 | Unrelated data | Voronoi diagrams, trilinear coordinates, heat maps |
| 2 | Hierarchical networks | Treemaps, Voronoi treemaps |
| 3 | Planar networks | Voronoi maps |
| 4 | Unstructured networks | Graphs, matrices |
| 5 | Combined networks | Chord diagrams, stacked models |
| Examples of Application Fields | Common Visualization Models (Topology Complexity Is Presented in Parentheses) | Alternative Visualization Models |
|---|---|---|
| Bioinformatics | Heat maps (1), graphs (4), planar networks (3), matrices (4), etc. | Treemaps (2), Voronoi diagrams (1), VR (5) |
| Sociology, economy | Hierarchical networks (2), unrelated data (1), matrices (4), etc. | Voronoi treemaps (2), VR (5), AR (5) |
| Agrosphere and soil science, geology | Geomaps (1), planar networks (3), combined networks (5), etc. | Trilinear coordinate model (1), Voronoi maps (3), VR (5) |
| Medicine and neuroscience | Graph of a function (4), trees (4), etc. | Chord diagrams (5) |
| Cyber-security | Trees (2), matrices (4), scatter plots (1), parallel coordinates (1), heat maps (1), etc. | Treemaps (2), Voronoi maps (3), stacked models (5) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolomeets, M.; Desnitsky, V.; Kotenko, I.; Chechulin, A. Graph Visualization: Alternative Models Inspired by Bioinformatics. Sensors 2023, 23, 3747. https://doi.org/10.3390/s23073747
Kolomeets M, Desnitsky V, Kotenko I, Chechulin A. Graph Visualization: Alternative Models Inspired by Bioinformatics. Sensors. 2023; 23(7):3747. https://doi.org/10.3390/s23073747
Chicago/Turabian StyleKolomeets, Maxim, Vasily Desnitsky, Igor Kotenko, and Andrey Chechulin. 2023. "Graph Visualization: Alternative Models Inspired by Bioinformatics" Sensors 23, no. 7: 3747. https://doi.org/10.3390/s23073747
APA StyleKolomeets, M., Desnitsky, V., Kotenko, I., & Chechulin, A. (2023). Graph Visualization: Alternative Models Inspired by Bioinformatics. Sensors, 23(7), 3747. https://doi.org/10.3390/s23073747











