3.2.1. Design Methodology and Considerations
In the previous section, we reviewed various conventional and neural network methods for epilepsy detection, along with their pros and cons. As reviewed earlier, both multiple- and single-channel approaches have been proposed. This depends on the targeted application. Most of these approaches that are high performance are multi-channel and computationally intensive. Neural network architectures generally involve deep architectures, while conventional approaches involve complex feature extraction stages.
Compact wearable devices, wireless sensor networks, and associated data processing are of increasing importance for diagnosis and patient monitoring. One should also consider minimizing the number of channels for the wearing comfort of ambulatory patients. Many hardware implementations have, therefore, adopted designs utilizing a low number of EEG channels and even a single channel. The application of such systems can be seen with reference to the extended diagnosis of epilepsy (generalized or focal seizures) and in monitoring the effectiveness of the treatment. Monitoring, recording, and marking seizure activity in one or a few channels in ambulatory patients at home can augment the status information obtained from the full-fledged EEG captured at the hospital. In such cases, the choice of these few monitoring locations will be the prerogative of the treating neurologists. This decision can be made by the neurologists after diagnosing the epilepsy type and observing abnormal wave complex activity in the EEG at different scalp locations.
The current focus of our work is to propose a computationally light and hardware-friendly approach that can analyze a single EEG channel with high accuracy to detect seizure activity. Most of the existing approaches employ multiple channels of EEG to extract a group feature vector and label the EEG epoch as seizure (ictel) or non-seizure (non-ictel) activity. Such an approach does not localize the evidence of the abnormal seizure activity to the channel level. Hence, a neurologist has to accomplish this on their own to accept or reject the automatic classification result. Furthermore, an approach based on processing a channel individually has a better potential to be scalable with respect to the number of channels, allowing its deployment in devices using one, few, or all EEG channels. For a multi-channel detection framework, this will mean processing the given channels separately as proposed and then assembling the individual detection results for a final global decision. This, however, is not discussed in this work. Furthermore, the scope of the current work is limited to achieve low algorithmic computational complexity but does not cover hardware implementation. This could be undertaken in the future.
As discussed earlier, AE models have been proposed in the recent literature [
9,
17] for epileptic seizure detection. Shallow AEs (involving a single layer) have been shown in earlier works to be successful in the generation of effective encoded samples (as features for classification) [
17]. They learn the essential representations of data from a number of sample sets [
17]. They have been extensively utilized in areas such as image de-noising [
43], feature learning [
44], and data modeling [
45], etc. For any considered dimension of the latent space, ref. [
45] reports that the best performance for unsupervised learning is obtained with AE for image databases.
This work experiments with a trainable hybrid approach involving a shallow AE and a conventional classifier. A shallow AE, instead of a deep neural architecture, is considered to provide an encoded representation of the input signal. A shallow AE brings a smaller number of parameters to train and store and are computationally efficient. Theoretical evidence about the fact that shallow AE models indeed can be used as a feature learning mechanism for a variety of data models has been proved in [
45]. A similar mechanism has been adopted in this work to develop our model taking into account our data. Conventional classifiers have been used to realize seizure detection or classification on this encoded representation of the input signal. An effective lower-dimensional encoded signal (smaller feature vector) in turns reduces the computational complexity and training effort along with the improvement in classification performance.
The proposed system involves a two-stage training process that involves training the AE first and then training the classifier. An encoded representation of the signal is trained in the first stage of the training process involving both the encoder and the decoder. The decoder reconstructs the signal to its original shape, and the objective of training is to minimize the reconstruction error. In the second stage, the trained encoded signal samples are used to train the classifier. Various conventional classifier models have been experimented on to see possible compromises in computational costs and classification performances suitable for different scenarios. The tested classifiers include kNN, SVM, DT, QDA, NB and softmax. The classification results of these models are discussed in the coming sections.
For determining the optimal design, multiple AE hidden sizes and different conventional classifier models have been experimented with. Analysis has been conducted on multiple hidden sizes to find the right balance for a shallow network that is large enough to capture the actual nature of the data and yet small enough to run faster as compared to other network architectures in the literature.
A completely training-based approach is suitable for both the ease of initial development and re-training for introducing patient specificity and adaptability to provide enhanced performance. The training or re-training is essentially more computationally intensive than test or in-operation-mode computation. However, as the algorithm training is usually done offline in the initial training phase or re-training phase, it is the computational effort in the testing (or operational) mode that matters most with respect to the minimization of energy and the computational time of the device. A detailed analysis and comparison of the computational cost in the testing phase of the model will be provided in the upcoming sections.
A short review of the theory of shallow AEs is provided as follows.
3.2.2. Shallow Autoencoder
An autoencoder is an unsupervised learning technique for neural networks that learns efficient data representations (encoding) by training the network to ignore signal “noise”. It learns to transform the data from a high-dimensional space to a lower-dimensional space. They work by encoding the data to a 1-D vector, which can then be decoded to reconstruct the original data. As compared to principal component analysis (PCA), autoencoder works for both linear and non-linear surfaces, whereas PCA only works for linear surfaces.
A shallow AE with a single hidden layer comprised of
n neurons in the input/output layer and
m hidden neurons is developed with
, as shown in
Figure 2. The encoder layer is evaluated by sharing the weights
and bias vectors
. The decoder layer reconstructs the signal with the weights
and bias vectors
. These weights and biases of the model are calculated for the signal reconstruction error instead of the classification results. The scaled conjugate gradient (SCG) method was designated to update these weights and bias values. Ref. [
44] averts the decoder bias, but we consider it an essential part of our model. The encoding process for an input EEG signal
is modeled as:
where
represents the activation function in the encoder neurons. The choice of the activation function usually varies with different data [
44]. However, linear activation aids in developing a system with a low computational cost as compared to non-linear functions. Therefore, we selected linear activations for the encoding and decoding processes. The saturating linear transfer function (Satlin) [
46] for the encoder activation function is given as:
The decoder layer reconstructs the encoded vector
to its original form, as follows:
The transfer function to compute the decoder layer’s output is a linear function (Purelin) . These functions are carried out on each sample of the signal with the mean square error (MSE) loss function. MSE is selected in the training phase of the AE to compare the original and reconstructed signal, and the AE model is trained until the loss function achieves its minimum possible value. Moreover, during the training phase, the regularization parameter is used to avoid model over-fitting. The encoded representation of the signal is achieved through encoding, and the reconstruction of the signal to its original form is achieved through the decoder. The decoding process is added to ensure the best possible encoded representation of the data.
3.2.3. Processing Architecture
Figure 2 depicts the basic processing architecture of the proposed methodology. The process starts with EEG data segmentation. EEG signal epochs of 1 to 30 s are usually adapted for different seizure detection methods in the literature [
47]. These durations are short enough to capture the dynamics of seizure activity while still being long enough to provide sufficient data for analysis. Shorter epochs in the stated range can enable the detection of seizure onset with low latency and can easily provide instances that are artifact- and noise-free. Thus, a 1 s non-overlapping EEG epoch is selected in this paper for accurate marking at fine granularity. This determines the system latency is 1 s as it detects the seizure for each second of EEG recording. An AE with a single hidden layer processes each 1 s EEG epoch to produce its encoded representation, as shown in
Figure 2. The input signal is encoded using the encoder layer to obtain an encoded vector. Once the AE is trained, the decoder layer is discarded, and only the encoder layer is applied for the encoded representation of the data, which is then used as an input to the classifier.
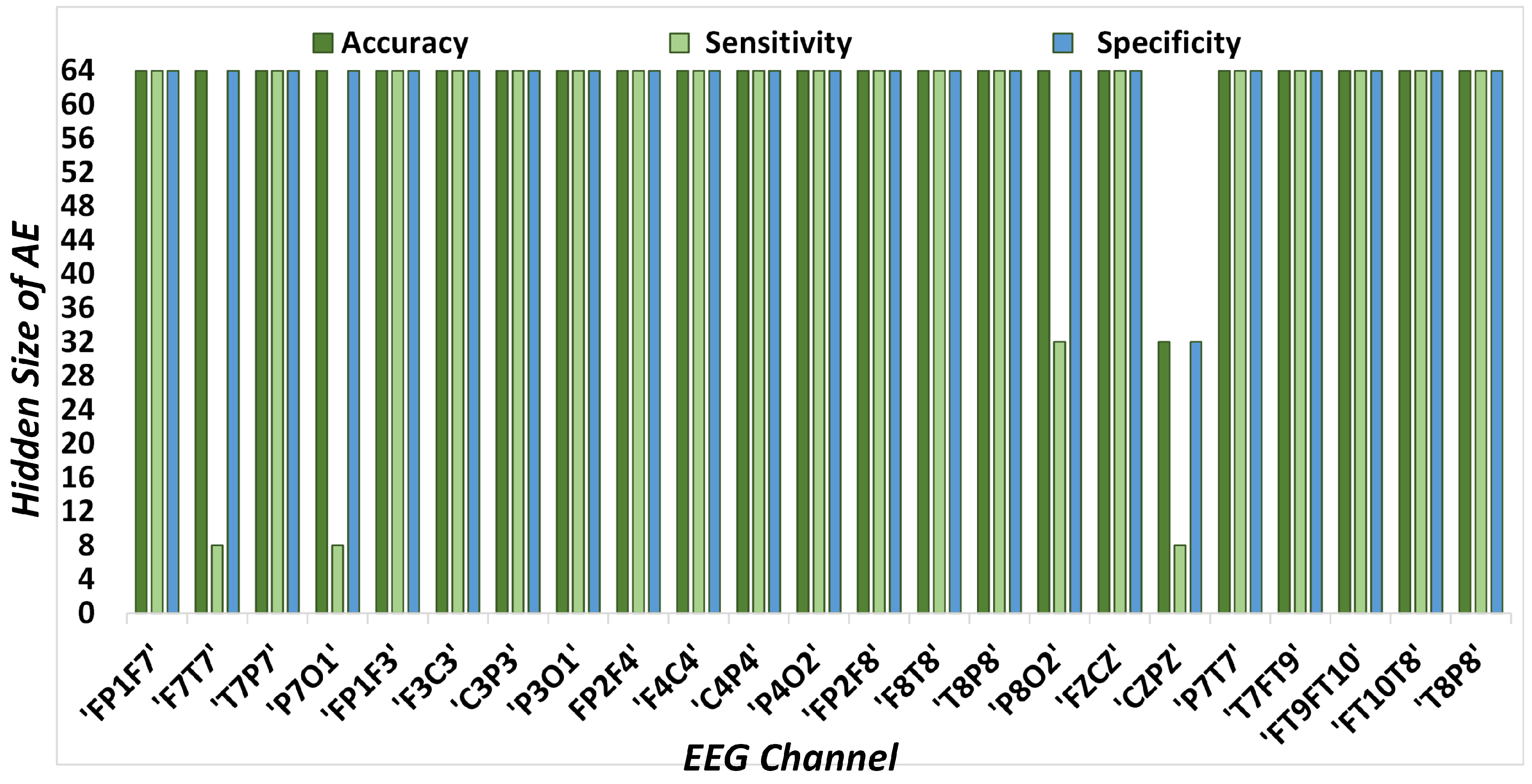
The steps necessary to process a 1 s EEG signal epoch are as follows. First, the database of our choice is at a sampling rate of 256 samples/s. Therefore, the input layer consists of a length of 256. The encoder compresses the input signal to 64 samples, whereas the decoder layer reconstructs the signal to its original length of 256, providing a compression ratio of 4 in our core experiment. Multiple experiments have been carried to determine the optimum value of the AE’s hidden size to obtain the best performance. However, for large values of the AE’s hidden size, the algorithm tends to learn very local features of the input data, which reduces the classification performance.
Table 1 encapsulates different lengths of encoded representations of EEG signals used in this paper, along with the corresponding data reduction ratios. The analysis of multiple AE hidden sizes is provided to evaluate the classification performance along with the computational complexity of the system. Therefore, the number of arithmetic operations (AO) for the AE and classifier are also presented to evaluate the exact computations with corresponding classification results. Our primary experiment deals with an AE hidden size 64, providing 75% diminution in the data. However, other levels are also part of our analysis and produce comparable results, even applying more than 90% data reduction. Consequently, the user has the choice to select any AE hidden size and achieve the corresponding data reduction among the listed options.
In order to categorize the data as epileptic or non-epileptic, the encoded representation of EEG epochs from the output of the encoder is used. Various machine learning algorithms are used for classification, such as kNN, SVM, DT, QDA, NB, and softmax layers. Adding a deep neural network (DNN) model may provide good results, but its computational cost would be quite high.
kNN: We selected 10 nearest neighbors according to their Euclidean distance for the kNN classifier. Weighted kNN is used to handle uncertain conditions with a squared inverse distance weight kernel. In order to encapsulate the uncertain outliers and to deal with incomplete and inconsistent information existing in the features, the exhaustive neutrosophic set was used to determine the decision criteria [
48].
SVM: For the SVM classifier, multiple kernel functions were tested. The impact of various kernel functions on SVM characteristics is highly variable. In this paper, we used the Gaussian kernel with random search. The hyper-parameters were randomly sampled from a specified distribution, and the model was trained and evaluated for each set of sampled hyper-parameters. An optimized hyperplane in kernel space was generated by a Gaussian function of kernel scale 2 and kernel offset 0.1. Training instances are separable for these parameters and yield optimal results. The feature space that training data is mapped to is determined by the kernel width. The optimal kernel scale value is the one that yields the best performance on the test set.
QDA: QDA is similar to LDA but without the assumption that the classes share the same covariance matrix, i.e., each class has its own covariance matrix. In this case, the boundary between classes is a quadratic surface instead of a hyperplane.
DT: DT is listed as the least computationally expensive classifier [
49]. To find the best split, we used the ’exact search’ algorithm with a tree of 159 nodes and the ’gdi’ split criterion.
NB: NB is based on Bayes’ theorem derived from the conditional probabilistic theory with an assumption of independence among predictors [
4]. In simple terms, an NB classifier assumes that the presence of a particular feature in a class is unrelated to the presence of any other feature. We used the Gaussian kernel function with a normal distribution for this classifier.
Softmax: Softmax layers are often used as the last layer of neural networks trained for data classification [
5]. We trained our softmax layer over an encoded vector in the same manner as previously listed classifiers. The ’binary cross-entropy’ function was used with the SCG algorithm to update the weight and bias values.