Self-Supervised Contrastive Learning for Medical Time Series: A Systematic Review
Abstract
1. Introduction
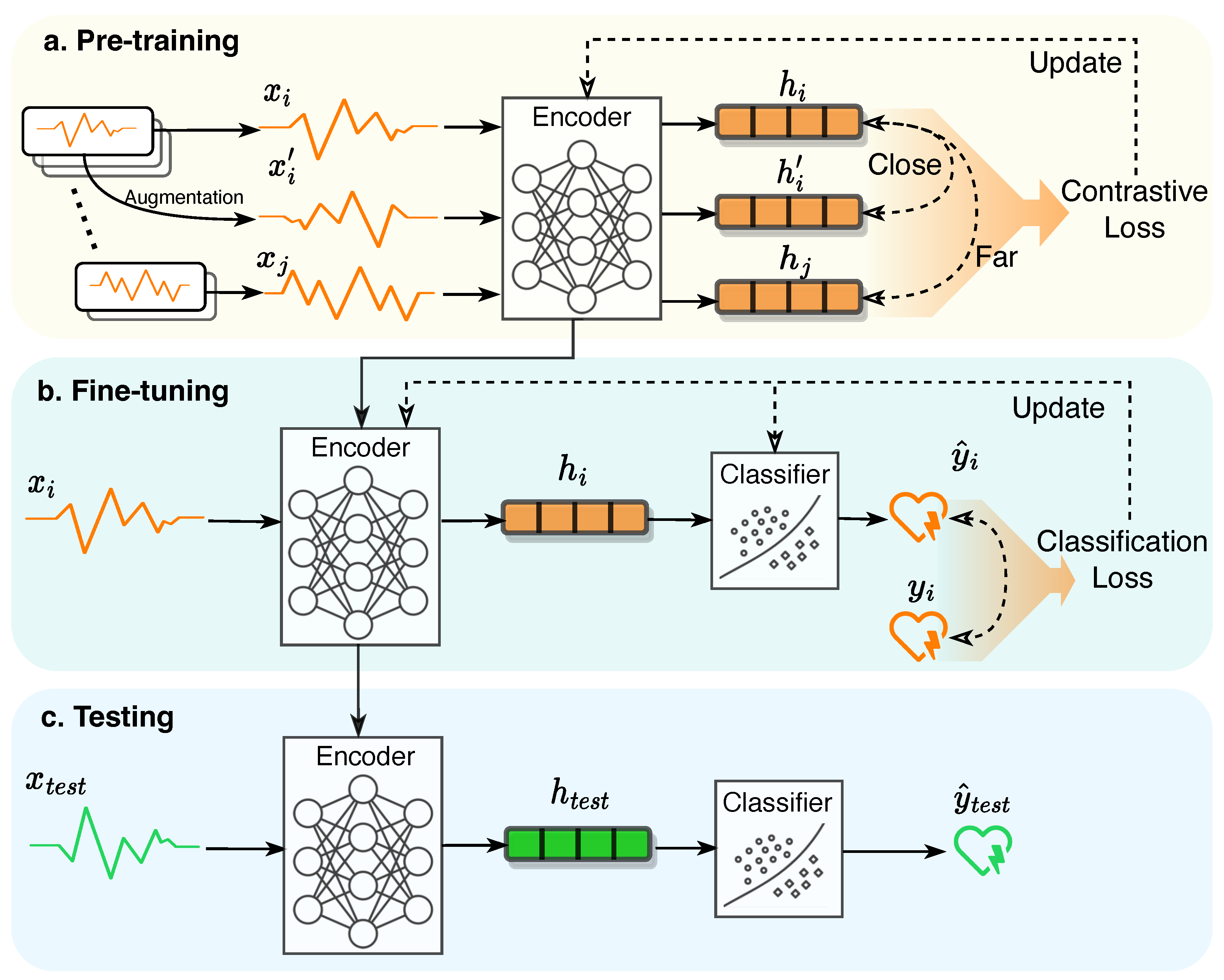
1.1. Self-Supervised Contrastive Learning
1.2. Systematic Review Objectives
- What studies have been conducted in the intersection of self-supervised contrastive learning and medical time series analysis? See Section 3.1.
- Which specific types of medical time series have been investigated in the literature mentioned above? See Section 3.2.
- What healthcare scenarios or applications are commonly observed in this scope? See Section 3.3.
- How are contrastive learning models designed in terms of sample augmentation, pretext task, encoder architecture, and contrastive loss functions? See Section 3.4, Section 3.5, Section 3.6 and Section 3.7.
- Which public datasets are commonly used, and what are their statistics? See Section 3.8.
- What are the current challenges and future directions in this field? See Section 4.
2. Methods
2.1. Databases and Search Strategy
2.2. Eligibility Criteria
3. Results
3.1. Overview
3.2. Data Types of Medical Time Series
| Study | Data Type | Application | Augmentation | Pretext Task | Pre-Training Encoder | Fine-Tuning Classifier | Contrastive Loss | Datasets | Transfer? | Code? |
|---|---|---|---|---|---|---|---|---|---|---|
| Sarkar et al. (2021) [56] | Abdominal ECG | Maternal and fetal stress detection | Jittering; Scaling; Flipping; Temporal inversion; Permutation; Time-warping | Augmentation type recognition | CNN | MLP | Cross entropy | AMIGOS; DREAMER; WESAD; SWELL; FELICITy | ✓ | ✓ |
| Jiang et al. (2021) [67] | Acceleration, Angular velocity | Parkinson’s Disease detection | Rotation and permutations | Predictive coding | CNN + GRU | One-class SVDD | MSE | mPower study | - | - |
| Song et al. (2021) [63] | Audio | Respiratory sound classification | Jittering; Time shifting; Time stretching; Pitch shifting | Contrastive mapping | CNN | Logistic regression | NT-Xent | ICBHI 2017 | - | - |
| Chen et al. (2022) [62] | Audio | COVID-19 detection | Block masking in time and frequency domains | Neighboring detection | CNN + Transformer | MLP | NT-Xent | DiCOVA - ICASSP 2022 | - | - |
| de Vries et al. (2022) [66] | CTG | fetal health detection | - | Predictive coding | GRU + MLP | - | Cosine distance + triplet loss | Dutch STAN trial; A healthy dataset | - | - |
| Mehari et al. (2022) [46] | ECG | Cardiac abnormalities diagnosis | Time out; Random resized crop; Jittering | Predictive coding | LSTM | Linear layer | InfoNCE loss | CinC 2020; Chapman; Ribeiro; PTB-XL | ✓ | - |
| Li et al. (2022) [47] | ECG | Cardiac abnormality detection | - | Contrastive mapping | BiLSTM-CNN; TimeGAN | CNN | NT-Xent | MIT-BIH; PTB | - | - |
| Kiyasseh et al. (2021) [18] | ECG | Cardiac arrhythmia classification | Time-wise and channel-wise neighboring | Contrastive mapping | CNN | Linear layer | NT-Xent | PhysioNet 2020; Chapman; PhysioNet 2017; Cardiology | ✓ | ✓ |
| Luo et al. (2021) [48] | ECG | Cardiac arrhythmia classification | Reorganization | Detect organization operation | CNN | - | Cross-entropy | PhysioNet 2017; CPSC 2018 | ✓ | - |
| Chen et al. (2021) [49] | ECG | Cardiac arrhythmia classification | Daubechies wavelet transform; Random crop | Contrastive mapping | ResNet + MLP | - | NT-Xent | PTB-XL; ICBEB 2018; PhysioNet 2017 | ✓ | - |
| Wei et al. (2022) [50] | ECG | Cardiac arrhythmia classification | Trials discrimination | Contrastive mapping | Causal CNN | Logistic regression | Multi-similartiy loss | MIT-BIH; Chapman; | - | - |
| Kiyasseh et al. (2021) [51] | ECG | Cardiac arrhythmia clustering; Sex and age clustering | - | Detect clinical prototype | CNN | Linear layer | NCE loss | Chapman; PTB-XL | - | - |
| Nguyen et al. (2020) [52] | ECG | Cardiac arrhythmia detection | - | Predictive coding | LSTM-based autoencoder | MLP | MSE + Cross entropy | MNIST; MIT-BIH | - | - |
| Yang et al. (2022) [53] | ECG | Cardiac events diagnostic | Frequency masking; Croping and resizing; R-peak masking; Channel masking | Momentum contrast | ResNet | MLP | NT-Xent + KL-divergence | CPSC 2018; | ✓ | |
| Mohsenvand et al. (2020) [28] | EEG | Emotion recognition; Seizure detection Sleep-stage scoring | Time shifting; Block masking; Amplitude scaling; Band-stop filtering; DC shifting; Jittering | Contrastive mapping | CNN | LSTM | NT-Xent | SEED dataset; TUH dataset; SleepEDF | - | - |
| He et al. (2022) [29] | EEG | Motor-Imagery classification | - | Predictive coding | CNN + LSTM | Linear layer | MSE | MI-2; BCIC IV 2a | - | - |
| Han et al. (2021) [71] | EEG | Motor-Imagery classification | Jittering; DC shift; Temporal roll, Amplitude scale, Temporal cutout, Crop and upsample | Contrastive mapping | CNN | CNN | NT-Xent | BCIC IV 2a | - | - |
| Wagh et al. (2021) [30] | ECG | EEG grade; Eye state; Demographics classification | Randomly flipping; Jittering | Hemipheric symmetry; Behavioral state estimation; Age contrastive | ResNet | - | Triplet loss | TUAB; MPI LEMON | - | ✓ |
| Xu et al. (2020) [20] | EEG | Seizure detection | Scaling transformations | Predicting transformation types | CNN | CNN | Cross-entropy | UPenn and Mayo Clinic’s seizure detection challenge | - | - |
| Ho et al. (2022) [31] | EEG | Seizure detection | Graph-based pair sampling | Contrastive mapping | GNN | Thresholding | NCE+MSE | TUSZ [72] | - | ✓ |
| Banville et al. (2019) [21] | EEG | Sleep scoring | Neighboring; Temporal shuffling | Contrastive mapping | CNN | Linear layer | Absolute distance | Sleep-EDF; MASS session3 | - | - |
| Yang et al. (2021) [32] | EEG | Sleep stage classification | Bandpass filtering; Jittering; Channel flipping | Contrastive mapping | STFT + CNN | Logistic regression | Triplet loss | SHHS, Sleep-EDF, MGH Sleep | - | ✓ |
| Xiao et al. (2021) [33] | EEG | Sleep stage classification | - | Predictive coding | CNN + LSTM | Linear layer | InfoNEC loss + Cross-entropy | Sleep-EDF; ISRUC | - | ✓ |
| Ye et al. (2022) [34] | EEG | Sleep stage classification | - | Predictive coding | ResNet + GRU | Linear layer | InfoNEC loss | Sleep-EDF; ISRUC | - | ✓ |
| Jiang et al. (2021) [35] | EEG | Sleep stage classification | Crop + resize; Permutation | Contrastive mapping | ResNet | MLP | NT-Xent | Sleep-EDF; Sleep-EDFx; Dod-O; Dod-H | - | ✓ |
| Cheng et al. (2020) [36] | EEG, ECG | Motor-Imagery classification; Cardiac arrhythmia classification | Block masking with noise | Contrastive mapping | ResNet | Logistic regression | InfoNCE | Physionet Motor Imagery; MIT-BIH | - | - |
| Ren et al. (2022) [37] | EEG, ECG | Sleep stage classification; Cardiac arrhythmia classification | - | Predictive coding | MLP | CNN | Cross entropy | Sleep-EDF; MIT-BIH-SUP | - | - |
| Huijben et al. (2022) [38] | EEG, EOG, EMG, Audio | Sleep stage clustering; Speakers clustering | - | Predictive coding | CNNs | SOM | InfoNCE | MASS; LibriSpeech | - | - |
| Saeed et al. (2021) [39] | EEG, EOG, Heart rate, GSR, Acceleration, Angular velocity | Activity recognition; Sleep stage scoring; Stress detection; WIFI sensing | Permutating; Channel shuffling; Timewarp; Scaling; Jittering; etc. | Blend detection; Augmentation type recognition; Feature prediction from masked window; etc. | CNN | Linear classifier | Huber loss; MSE; Triplet loss; Cross-entropy | HHAR; MobiAct; UCI HAR; HAPT; Sleep-EDF; etc. | - | - |
| Zhang et al. (2022) [40] | EEG, ECG | Sleep disorder classification; Seizure detection | Jittering; Frequency masking; Time masking | Contrastive mapping | CNN | MLP/KNN | NT-Xent | SleepEDF; Eplipsy Seizure; HAR, etc. | ✓ | ✓ |
| Chen et al. (2021) [58] | ICU | Forecast adverse surgical events | - | Predictive coding | LSTM | LSTM | Cross-entropy | OR dataset; MIMIC dataset | ✓ | ✓ |
| Weatherhead et al. (2022) [60] | ICU | Mortality prediction; Diagnostic group classification; Circulatory failure prediction; Cardio pulmonary arrest prediction | Neighboring | Detect neighbors | Dilated causal CNN | LSTM | Min-max GAN loss | HiRID dataset; High-frequency ICU | - | |
| Manduchi et al. (2021) [59] | ICU | Patient health state tracking | - | Predictive coding | VAE-LSTM | - | KL-divergence; Cross-entropy | MNIST; Fashion-MNIST; eICU dataset | - | ✓ |
| Ballas et al. (2022) [57] | PCG | Heart sound classification | High-pass filtering; Jittering + upsampling | Contrastive mapping | CNN | MLP | NT-Xent | PhysioNet 2016; PhysioNet 2022 | - | |
| Zhao et al. (2020) [65] | Acceleration, Heart rate, Bioradar, etc. | Sleep stage classification; Insomnia detection | Rotation | Rotation degrees recognition | CNN | Bi-LSTM- CRF | Cross-entropy | Sleep Bioradiolocation; PSG dataset; etc. | - | - |
| Study | Data type | Applications | Model | Classifier | Loss | Datasets | Transfer? | Code? | Notes |
|---|---|---|---|---|---|---|---|---|---|
| Gedon et al. (2022) [54] | ECG | Cardiac abnormality detection | ResNet | Linear layer | MSE | Train: CODE; Test: CPSC 2018, PTB-XL | ✓ | - | Reconstruct the masked signal |
| Lee et al. (2021) [55] | ECG | Cardiac arrhythmia classification | ResNet | MLP | - | CPSC, PT-BXL, Chapman-Shaoxing | ✓ | - | - |
| Spathis et al. (2021) [64] | Hear rate, Acceleration | Personalized health-related outcomes prediction | CNN and GRU | Logistic regression | MSE; Quantile loss | The Fenland study | ✓ | ✓ | Sequence to sequence mapping |
| Tang et al. (2022) [41] | EEG | Seizure detection | DCGRUs | MLP | - | TUSZ, In-house dataset | - | ✓ | Forecast the future sequence |
| Yang et al. (2022) [42] | EEG | Seizure detection and forecasting | Conv-LSTM | MLP | Cross entropy | TUH seizure, EPILEPSIAE dataset, RPAH dataset (private) | - | ✓ | - |
| Stuldreher et al. (2022) [43] | EEG, EDA, ECG | Attentional engagement state clustering | PCoA+UMAP | Kmeans | - | Physiological Synchrony Selective Attention | - | - | Clustering |
| Wever et al. (2021) [69] | ICU, Menstrual tracking data | Mortality prediction, Discontinuation of birth control methods prediction | GRU-Decay | MLP | MaskedMSE | Physionet challenge 2012, Clue app data | - | ✓ | - |
| Edinburgh et al. (2020) [61] | ICU | Artefact detection on ICU physiological data | CNN-based VAE | - | MSE | Not mentioned (ABP waveform data from single anonymized patient throughout a stay) | - | ✓ | Reconstruct the signal |
3.3. Medical Applications
3.4. Augmentations
3.4.1. Overview of Data Augmentation in Time Series
3.4.2. Transforming Augmentation
3.4.3. Masking Augmentation
3.4.4. Neighboring Augmentation
3.5. Pretext Tasks
3.6. Model Architecture
3.6.1. Pre-Training Encoder
3.6.2. Fine-Tuning Classifier
3.7. Contrastive Loss
3.8. Public Datasets
| Dataset | Data Type | Subjects | Frequency (Hz) | Channels | Task |
|---|---|---|---|---|---|
| MotionSense [92] | Acceleration, Angular velocity | 24 | 50 | 12 | Activity Recognition |
| HHAR [93] | Acceleration, Angular velocity | 9 | 50–200 | 16 | Activity Recognition |
| MobiAct [94] | Acceleration, Angular velocity | 57 | 20 | 6 | Activity Recognition |
| UCI HAR [95] | Acceleration, Angular velocity | 30 | 50 | 6 | Activity Recognition |
| PSG dataset [96] | Acceleration, HR, Steps | 31 | - | Acc.: 3 HR: 1 Steps: 1 | Sleep study |
| Dutch STAN trial [97] | CTG | 5681 | - | - | Fetal monitoring |
| DiCOVA-ICASSP 2022 challenge [98] | Cough, Speech, Breath | - | 44.1 k | - | COVID detection |
| CODE [99] | ECG | 1,558,415 | 300–1000 | 12 | ECG abnormalities detection |
| Ribeiro [100] | ECG | 827 | 12 | Automatic diagnosis of ECG | |
| PhysioNet 2020 [101] | ECG | 6877 | 12 | ECG classification | |
| MIT-BIH Arrhythmia [102] | ECG | 47 | 125 | 2 | Cardiac arrhythmia study |
| PhysioNet 2017 [103] | ECG | 8528 (recordings) | 300 | 1 | AF(ECG) Classification |
| CPSC 2018 (ICBEB2018) [104] | ECG | 6877 | 500 | 12 | Heart diseases study |
| PTB [105] | ECG | 290 | 125 | 14 | Heart diseases study |
| Chapman-Shaoxing [106] | ECG | 10,646 | 500 | 12 | Cardiac arrhythmia study |
| Cardiology [107] | ECG | 328 | 1 | Arrhythmia detection and classification | |
| PTB-XL [108] | ECG | 18,869 | 100 | 12 | Heart diseases study |
| MIT-BIH-SUP [109] | ECG | 78 | 128 | - | Supplement of supraventricular arrhythmias in MIT-BIH |
| Physiological Synchrony Selective Attention [110] | ECG, EEG, EDA | 26 | 1024 | EEG:32 EDA: 2 ECG:2 | Attention focus study |
| SHHS [111,112] | ECG, EEG, EOG, EMG, SpO2, RR | 9736 | EEG: 125 EOG: 50 EMG: 125 ECG: 125/250 SpO2: 1 RR: 10 | EEG: 2 EOG: 2 EMG: 1 ECG: 1 SpO2: 1 RR: 1 | Sleep-disordered breathing study |
| AMIGOS [113] | ECG, EEG, GSR | 40 | ECG: 256 EEG: 128 GSR: 128 | ECG: 4 EEG: 14 GSR: 2 | Emotional states recognition |
| MPI LEMON [114] | EEG, fMRI | 227 | EEG: 2500 | EEG: 62 | Mind-body-emotion interactions study |
| PhysioNet 2016 [73] | ECG, PCG | 3126 | 2000 | 2 | Heart Sound Recordings classification |
| WESAD [115] | ECG, Acceleration, etc. | 15 | ECG: 700 | ECG: 1 | Wearable Stress and Affect Detection |
| SWELL [116] | ECG, Facial expressions, etc. | 25 | ECG: 2048 | - | Work psychology study |
| The Fenland study [117] | ECG, HR, Acceleration, etc. | 2100 | - | - | Obesity, type 2 diabetes, and related metabolic disorders study |
| SEED [118,119] | EEG | 15 | 200 | 62 | Emotion Recognition |
| TUSZ [72] | EEG | 315 | 250 | 21 | Seizure study |
| TUAB [120] | EEG | 564 | 250 | 21 | ECG abnormalities study |
| EEG Motor Movement/ Imagery [121] | EEG | 109 | 160 | 64 | Motor-Imagery classification |
| BCI Competition IV-2A [122] | EEG | 9 | 250 | 22 | Motor-Imagery classification |
| Sleep-EDFx [123,124] | EEG | 197 | 100 | 2 | Sleep study |
| MGH Sleep [111] | EEG | 2621 | 200 | 6 | Sleep study |
| MI-2 Dataset [125] | EEG | 25 | 200 | 62 | Motor-Imagery classification |
| EPILEPSIAE [126] | EEG | 275 | 250 | - | Seizure study |
| UPenn Mayo Clinic’s Seizure Detection Challenge [127] | EEG (Intracranial) | 4 dogs 8 human | 400 | 16 | Seizure study |
| DOD-H [128] | EEG (PSG data) | 25 | 250 | 12 | Sleep study |
| DOD-O [128] | EEG (PSG data) | 55 | 250 | 8 | Sleep study |
| DREAMER [129] | EEG, ECG | 23 | ECG: 256 | ECG: 4 | Affect recognition |
| MASS [90] | EEG, EMG, EOG | 200 | 256 | 16–21 * | Sleep study |
| PhysioNet 2018 [130] | EEG, EOG, EMG, ECG, SaO2 | 1985 | 200 | 5 | Diagnosis of sleep disorders |
| ISRUC-SLEEP [91] | EEG, EOG, EMG, ECG, SaO2 | 100/8/10 * | EEG: 200 EOG: 200 EMG: 200 ECG: 200 SaO2: 12.5 | EEG: 6 EOG: 2 EMG: 2 ECG: 1 SaO2: 1 | Sleep study |
| Sleep-EDF [123,124] | EEG, EOG, chin EMG | 20 | 100 | 2 | |
| MIT DriverDb [68] | ECG, EMG, EDA, PR | 17 | ECG: 496 EMG: 15.5 EDA: 31 PR: 31 | ECG: 1 EMG: 1 EDA: 1 PR: 1 | Stress detection |
| HiRID [79,131] | ICU | 33,000+ | - | ||
| eICU [80] | ICU | - | - | 160 variables | - |
| PhysioNet 2012 [82] | ICU | 12,000 | - | 37 | Mortality prediction |
| MIMIC-III [78] | ICU | 4000+ | - | - | - |
| PhysioNet 2022 [124,132,133] | PCG | 1568 | 4000 | 5 | Heart Murmur Detection |
| ICBHI 2017 [134] | Respiratory sound | 126 | 4000 | 1 | Computational lung auscultation |
| LibriSpeech dataset [135] | Voice | 251 | 16k | - | Speech Recognition |
| mPower data [77] | Voice, Walking kinematics | Walking: 3101 | - | - | Parkinson disease study though mobile data |
3.9. Model Transferability and Code Availability
3.10. Evaluation Metrics
4. Discussion and Opening Challenges
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Spathis, D.; Perez-Pozuelo, I.; Brage, S.; Wareham, N.J.; Mascolo, C. Learning generalizable physiological representations from large-scale wearable data. arXiv 2020, arXiv:2011.04601. [Google Scholar]
- Che, Z.; Cheng, Y.; Zhai, S.; Sun, Z.; Liu, Y. Boosting deep learning risk prediction with generative adversarial networks for electronic health records. In Proceedings of the 2017 IEEE International Conference on Data Mining (ICDM), New Orleans, LA, USA, 18–21 November 2017; pp. 787–792. [Google Scholar]
- Cornet, V.P.; Holden, R.J. Systematic review of smartphone-based passive sensing for health and wellbeing. J. Biomed. Inform. 2018, 77, 120–132. [Google Scholar] [CrossRef] [PubMed]
- Morid, M.A.; Sheng, O.R.L.; Dunbar, J. Time series prediction using deep learning methods in healthcare. ACM Trans. Manag. Inf. Syst. 2023, 14, 1–29. [Google Scholar] [CrossRef]
- Harutyunyan, H.; Khachatrian, H.; Kale, D.C.; Ver Steeg, G.; Galstyan, A. Multitask learning and benchmarking with clinical time series data. Sci. Data 2019, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Shurrab, S.; Duwairi, R. Self-supervised learning methods and applications in medical imaging analysis: A survey. PeerJ Comput. Sci. 2022, 8, e1045. [Google Scholar] [CrossRef]
- Chowdhury, A.; Rosenthal, J.; Waring, J.; Umeton, R. Applying self-supervised learning to medicine: Review of the state of the art and medical implementations. Informatics 2021, 8, 59. [Google Scholar] [CrossRef]
- Pan, L.; Feng, Z.; Peng, S. A review of machine learning approaches, challenges and prospects for computational tumor pathology. arXiv 2022, arXiv:2206.01728. [Google Scholar]
- Wang, P.; Li, Y.; Reddy, C.K. Machine learning for survival analysis: A survey. ACM Comput. Surv. (CSUR) 2019, 51, 1–36. [Google Scholar] [CrossRef]
- Shickel, B.; Tighe, P.J.; Bihorac, A.; Rashidi, P. Deep EHR: A survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J. Biomed. Health Inform. 2017, 22, 1589–1604. [Google Scholar] [CrossRef]
- Faust, O.; Hagiwara, Y.; Hong, T.J.; Lih, O.S.; Acharya, U.R. Deep learning for healthcare applications based on physiological signals: A review. Comput. Methods Programs Biomed. 2018, 161, 1–13. [Google Scholar] [CrossRef]
- Hasan, N.I.; Bhattacharjee, A. Deep learning approach to cardiovascular disease classification employing modified ECG signal from empirical mode decomposition. Biomed. Signal Process. Control 2019, 52, 128–140. [Google Scholar] [CrossRef]
- Chen, H.; Song, Y.; Li, X. A deep learning framework for identifying children with ADHD using an EEG-based brain network. Neurocomputing 2019, 356, 83–96. [Google Scholar] [CrossRef]
- Baker, S.; Xiang, W.; Atkinson, I. Continuous and automatic mortality risk prediction using vital signs in the intensive care unit: A hybrid neural network approach. Sci. Rep. 2020, 10, 21282. [Google Scholar] [CrossRef] [PubMed]
- Wickramaratne, S.D.; Mahmud, M.S. Bi-directional gated recurrent unit based ensemble model for the early detection of sepsis. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 70–73. [Google Scholar]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A simple framework for contrastive learning of visual representations. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual, 13–18 July 2020; pp. 1597–1607. [Google Scholar]
- Liu, X.; Zhang, F.; Hou, Z.; Mian, L.; Wang, Z.; Zhang, J.; Tang, J. Self-supervised learning: Generative or contrastive. IEEE Trans. Knowl. Data Eng. 2021, 35, 857–876. [Google Scholar] [CrossRef]
- Kiyasseh, D.; Zhu, T.; Clifton, D.A. Clocs: Contrastive learning of cardiac signals across space, time, and patients. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual, 18–24 July 2021; pp. 5606–5615. [Google Scholar]
- You, Y.; Chen, T.; Sui, Y.; Chen, T.; Wang, Z.; Shen, Y. Graph contrastive learning with augmentations. Adv. Neural Inf. Process. Syst. 2020, 33, 5812–5823. [Google Scholar]
- Xu, J.; Zheng, Y.; Mao, Y.; Wang, R.; Zheng, W.S. Anomaly detection on electroencephalography with self-supervised learning. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Virtual, 16–19 December 2020; pp. 363–368. [Google Scholar]
- Banville, H.; Albuquerque, I.; Hyvärinen, A.; Moffat, G.; Engemann, D.A.; Gramfort, A. Self-supervised representation learning from electroencephalography signals. In Proceedings of the 2019 IEEE 29th International Workshop on Machine Learning for Signal Processing (MLSP), Pittsburgh, PA, USA, 13–16 October 2019; pp. 1–6. [Google Scholar]
- Spathis, D.; Perez-Pozuelo, I.; Marques-Fernandez, L.; Mascolo, C. Breaking away from labels: The promise of self-supervised machine learning in intelligent health. Patterns 2022, 3, 100410. [Google Scholar] [CrossRef]
- Iwana, B.K.; Uchida, S. An empirical survey of data augmentation for time series classification with neural networks. PLoS ONE 2021, 16, e0254841. [Google Scholar] [CrossRef]
- Yu, K.H.; Beam, A.L.; Kohane, I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018, 2, 719–731. [Google Scholar] [CrossRef]
- Albelwi, S. Survey on Self-Supervised Learning: Auxiliary Pretext Tasks and Contrastive Learning Methods in Imaging. Entropy 2022, 24, 551. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Syst. Rev. 2021, 372, n71. [Google Scholar]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Mohsenvand, M.N.; Izadi, M.R.; Maes, P. Contrastive representation learning for electroencephalogram classification. In Proceedings of the Machine Learning for Health, PMLR, Virtual, 7–8 August 2020; pp. 238–253. [Google Scholar]
- He, Y.; Lu, Z.; Wang, J.; Ying, S.; Shi, J. A Self-Supervised Learning Based Channel Attention MLP-Mixer Network for Motor Imagery Decoding. IEEE Trans. Neural Syst. Rehabil. Eng. 2022, 30, 2406–2417. [Google Scholar] [CrossRef] [PubMed]
- Wagh, N.; Wei, J.; Rawal, S.; Berry, B.; Barnard, L.; Brinkmann, B.; Worrell, G.; Jones, D.; Varatharajah, Y. Domain-guided Self-supervision of EEG Data Improves Downstream Classification Performance and Generalizability. In Proceedings of the Machine Learning for Health, PMLR, Virtual, 6–7 August 2021; pp. 130–142. [Google Scholar]
- Ho, T.K.K.; Armanfard, N. Self-Supervised Learning for Anomalous Channel Detection in EEG Graphs: Application to Seizure Analysis. arXiv 2022, arXiv:2208.07448. [Google Scholar]
- Yang, C.; Xiao, D.; Westover, M.B.; Sun, J. Self-supervised eeg representation learning for automatic sleep staging. arXiv 2021, arXiv:2110.15278. [Google Scholar]
- Xiao, Q.; Wang, J.; Ye, J.; Zhang, H.; Bu, Y.; Zhang, Y.; Wu, H. Self-supervised learning for sleep stage classification with predictive and discriminative contrastive coding. In Proceedings of the ICASSP 2021-2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 1290–1294. [Google Scholar]
- Ye, J.; Xiao, Q.; Wang, J.; Zhang, H.; Deng, J.; Lin, Y. CoSleep: A multi-view representation learning framework for self-supervised learning of sleep stage classification. IEEE Signal Process. Lett. 2021, 29, 189–193. [Google Scholar] [CrossRef]
- Jiang, X.; Zhao, J.; Du, B.; Yuan, Z. Self-supervised contrastive learning for eeg-based sleep staging. In Proceedings of the IEEE 2021 International Joint Conference on Neural Networks (IJCNN), Virtual, 18–22 July 2021; pp. 1–8. [Google Scholar]
- Cheng, J.Y.; Goh, H.; Dogrusoz, K.; Tuzel, O.; Azemi, E. Subject-aware contrastive learning for biosignals. arXiv 2020, arXiv:2007.04871. [Google Scholar]
- Ren, C.; Sun, L.; Peng, D. A Contrastive Predictive Coding-Based Classification Framework for Healthcare Sensor Data. J. Healthc. Eng. 2022, 2022. [Google Scholar] [CrossRef] [PubMed]
- Huijben, I.A.; Nijdam, A.A.; Overeem, S.; van Gilst, M.M.; van Sloun, R.J. SOM-CPC: Unsupervised Contrastive Learning with Self-Organizing Maps for Structured Representations of High-Rate Time Series. arXiv 2022, arXiv:2205.15875. [Google Scholar]
- Saeed, A.; Ungureanu, V.; Gfeller, B. Sense and learn: Self-supervision for omnipresent sensors. Mach. Learn. Appl. 2021, 6, 100152. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, Z.; Tsiligkaridis, T.; Zitnik, M. Self-supervised contrastive pre-training for time series via time-frequency consistency. arXiv 2022, arXiv:2206.08496. [Google Scholar]
- Tang, S.; Dunnmon, J.; Saab, K.K.; Zhang, X.; Huang, Q.; Dubost, F.; Rubin, D.; Lee-Messer, C. Self-Supervised Graph Neural Networks for Improved Electroencephalographic Seizure Analysis. In Proceedings of the International Conference on Learning Representations, Virtual, 25–29 April 2022. [Google Scholar]
- Yang, Y.; Truong, N.D.; Eshraghian, J.K.; Nikpour, A.; Kavehei, O. Weak self-supervised learning for seizure forecasting: A feasibility study. R. Soc. Open Sci. 2022, 9, 220374. [Google Scholar] [CrossRef] [PubMed]
- Stuldreher, I.V.; Merasli, A.; Thammasan, N.; Van Erp, J.B.; Brouwer, A.M. Unsupervised Clustering of Individuals Sharing Selective Attentional Focus Using Physiological Synchrony. Front. Neuroergonomics 2022, 2, 750248. [Google Scholar] [CrossRef]
- Jackson, A.F.; Bolger, D.J. The neurophysiological bases of EEG and EEG measurement: A review for the rest of us. Psychophysiology 2014, 51, 1061–1071. [Google Scholar] [CrossRef] [PubMed]
- Craik, A.; He, Y.; Contreras-Vidal, J.L. Deep learning for electroencephalogram (EEG) classification tasks: A review. J. Neural Eng. 2019, 16, 031001. [Google Scholar] [CrossRef]
- Mehari, T.; Strodthoff, N. Self-supervised representation learning from 12-lead ECG data. Comput. Biol. Med. 2022, 141, 105114. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chang, H.; Jiang, M.; Su, Y. A Contrastive Learning Framework for ECG Anomaly Detection. In Proceedings of the IEEE 2022 7th International Conference on Intelligent Computing and Signal Processing (ICSP), Xi’an, China, 15–17 April 2022; pp. 673–677. [Google Scholar]
- Luo, C.; Wang, G.; Ding, Z.; Chen, H.; Yang, F. Segment Origin Prediction: A Self-supervised Learning Method for Electrocardiogram Arrhythmia Classification. In Proceedings of the 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 1–5 November 2021; pp. 1132–1135. [Google Scholar]
- Chen, H.; Wang, G.; Zhang, G.; Zhang, P.; Yang, H. CLECG: A Novel Contrastive Learning Framework for Electrocardiogram Arrhythmia Classification. IEEE Signal Process. Lett. 2021, 28, 1993–1997. [Google Scholar] [CrossRef]
- Wei, C.T.; Hsieh, M.E.; Liu, C.L.; Tseng, V.S. Contrastive Heartbeats: Contrastive Learning for Self-Supervised ECG Representation and Phenotyping. In Proceedings of the ICASSP 2022-2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Singapore, 22–27 May 2022; pp. 1126–1130. [Google Scholar]
- Kiyasseh, D.; Zhu, T.; Clifton, D. CROCS: Clustering and Retrieval of Cardiac Signals Based on Patient Disease Class, Sex, and Age. Adv. Neural Inf. Process. Syst. 2021, 34, 15557–15569. [Google Scholar]
- Nguyen, D.; Nguyen, P.; Do, K.; Rana, S.; Gupta, S.; Tran, T. Unsupervised Anomaly Detection on Temporal Multiway Data. In Proceedings of the 2020 IEEE Symposium Series on Computational Intelligence (SSCI), Canberra, ACT, Australia, 1–4 December 2020; pp. 1059–1066. [Google Scholar]
- Yang, W.; Feng, Q.; Lai, J.; Tan, H.; Wang, J.; Ji, L.; Guo, J.; Han, B.; Shi, Y. Practical Cardiac Events Intelligent Diagnostic Algorithm for Wearable 12-Lead ECG via Self-Supervised Learning on Large-Scale Dataset. 2022. Available online: https://www.researchsquare.com/article/rs-1796360/v1 (accessed on 3 February 2023).
- Gedon, D.; Ribeiro, A.H.; Wahlström, N.; Schön, T.B. First Steps Towards Self-Supervised Pretraining of the 12-Lead ECG. In Proceedings of the IEEE 2021 Computing in Cardiology (CinC), Brno, Czech Republic, 13–15 September 2021; Volume 48, pp. 1–4. [Google Scholar]
- Lee, B.T.; Kong, S.T.; Song, Y.; Lee, Y. Self-Supervised Learning with Electrocardiogram Delineation for Arrhythmia Detection. In Proceedings of the 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 1–5 November 2021; pp. 591–594. [Google Scholar]
- Sarkar, P.; Lobmaier, S.; Fabre, B.; González, D.; Mueller, A.; Frasch, M.G.; Antonelli, M.C.; Etemad, A. Detection of maternal and fetal stress from the electrocardiogram with self-supervised representation learning. Sci. Rep. 2021, 11, 24146. [Google Scholar] [CrossRef]
- Ballas, A.; Papapanagiotou, V.; Delopoulos, A.; Diou, C. Listen2YourHeart: A Self-Supervised Approach for Detecting Murmur in Heart-Beat Sounds. arXiv 2022, arXiv:2208.14845. [Google Scholar]
- Chen, H.; Lundberg, S.M.; Erion, G.; Kim, J.H.; Lee, S.I. Forecasting adverse surgical events using self-supervised transfer learning for physiological signals. NPJ Digit. Med. 2021, 4, 167. [Google Scholar] [CrossRef]
- Manduchi, L.; Hüser, M.; Faltys, M.; Vogt, J.; Rätsch, G.; Fortuin, V. T-dpsom: An interpretable clustering method for unsupervised learning of patient health states. In Proceedings of the Conference on Health, Inference, and Learning, Virtual, 8–10 April 2021; pp. 236–245. [Google Scholar]
- Weatherhead, A.; Greer, R.; Moga, M.A.; Mazwi, M.; Eytan, D.; Goldenberg, A.; Tonekaboni, S. Learning Unsupervised Representations for ICU Timeseries. In Proceedings of the Conference on Health, Inference, and Learning, PMLR, Virtual, 7–8 April 2022; pp. 152–168. [Google Scholar]
- Edinburgh, T.; Smielewski, P.; Czosnyka, M.; Cabeleira, M.; Eglen, S.J.; Ercole, A. DeepClean: Self-Supervised Artefact Rejection for Intensive Care Waveform Data Using Deep Generative Learning. In Intracranial Pressure and Neuromonitoring XVII; Springer: Berlin/Heidelberg, Germany, 2021; pp. 235–241. [Google Scholar]
- Chen, X.Y.; Zhu, Q.S.; Zhang, J.; Dai, L.R. Supervised and Self-supervised Pretraining Based COVID-19 Detection Using Acoustic Breathing/Cough/Speech Signals. In Proceedings of the ICASSP 2022–2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Singapore, 22–27 May 2022; pp. 561–565. [Google Scholar]
- Song, W.; Han, J.; Song, H. Contrastive embeddind learning method for respiratory sound classification. In Proceedings of the ICASSP 2021–2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 1275–1279. [Google Scholar]
- Spathis, D.; Perez-Pozuelo, I.; Brage, S.; Wareham, N.J.; Mascolo, C. Self-supervised transfer learning of physiological representations from free-living wearable data. In Proceedings of the Conference on Health, Inference, and Learning, Virtual, 8–10 April 2021; pp. 69–78. [Google Scholar]
- Zhao, A.; Dong, J.; Zhou, H. Self-supervised learning from multi-sensor data for sleep recognition. IEEE Access 2020, 8, 93907–93921. [Google Scholar] [CrossRef]
- de Vries, I.R.; Huijben, I.A.; Kok, R.D.; van Sloun, R.J.; Vullings, R. Contrastive Predictive Coding for Anomaly Detection of Fetal Health from the Cardiotocogram. In Proceedings of the ICASSP 2022–2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Singapore, 22–27 May 2022; pp. 3473–3477. [Google Scholar]
- Jiang, H.; Lim, W.Y.B.; Ng, J.S.; Wang, Y.; Chi, Y.; Miao, C. Towards parkinson’s disease prognosis using self-supervised learning and anomaly detection. In Proceedings of the ICASSP 2021–2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 3960–3964. [Google Scholar]
- Healey, J.A.; Picard, R.W. Detecting stress during real-world driving tasks using physiological sensors. IEEE Trans. Intell. Transp. Syst. 2005, 6, 156–166. [Google Scholar] [CrossRef]
- Wever, F.; Keller, T.A.; Symul, L.; Garcia, V. As easy as APC: Overcoming missing data and class imbalance in time series with self-supervised learning. arXiv 2021, arXiv:2106.15577. [Google Scholar]
- BioWink GmbH. Clue. 2020. Available online: https://helloclue.com/ (accessed on 15 January 2021).
- Han, J.; Gu, X.; Lo, B. Semi-supervised contrastive learning for generalizable motor imagery eeg classification. In Proceedings of the 2021 IEEE 17th International Conference on Wearable and Implantable Body Sensor Networks (BSN), Athens, Greece, 27–30 July 2021; pp. 1–4. [Google Scholar]
- Shah, V.; Von Weltin, E.; Lopez, S.; McHugh, J.R.; Veloso, L.; Golmohammadi, M.; Obeid, I.; Picone, J. The temple university hospital seizure detection corpus. Front. Neuroinformatics 2018, 12, 83. [Google Scholar] [CrossRef]
- Liu, C.; Springer, D.; Moody, B.; Silva, I.; Johnson, A.; Samieinasab, M.; Sameni, R.; Mark, R.; Clifford, G.D. Classification of Heart Sound Recordings-The PhysioNet Computing in Cardiology Challenge 2016. PhysioNet 2016. Available online: https://www.physionet.org/content/challenge-2016/1.0.0/papers/ (accessed on 4 March 2016).
- Wang, H.; Lin, G.; Li, Y.; Zhang, X.; Xu, W.; Wang, X.; Han, D. Automatic Sleep Stage Classification of Children with Sleep-Disordered Breathing Using the Modularized Network. Nat. Sci. Sleep 2021, 13, 2101–2112. [Google Scholar] [CrossRef]
- Tataraidze, A.; Korostovtseva, L.; Anishchenko, L.; Bochkarev, M.; Sviryaev, Y.; Ivashov, S. Bioradiolocation-based sleep stage classification. In Proceedings of the EMBC, Orlando, FL, USA, 16–20 August 2016; pp. 2839–2842. [Google Scholar]
- Harati, A.; Lopez, S.; Obeid, I.; Picone, J.; Jacobson, M.; Tobochnik, S. The TUH EEG CORPUS: A big data resource for automated EEG interpretation. In Proceedings of the 2014 IEEE Signal Processing in Medicine and Biology Symposium (SPMB), Philadelphia, PA, USA, 13 December 2014; pp. 1–5. [Google Scholar]
- Bot, B.M.; Suver, C.; Neto, E.C.; Kellen, M.; Klein, A.; Bare, C.; Doerr, M.; Pratap, A.; Wilbanks, J.; Dorsey, E.; et al. The mPower study, Parkinson disease mobile data collected using ResearchKit. Sci. Data 2016, 3, 160011. [Google Scholar] [CrossRef]
- Johnson, A.E.; Pollard, T.J.; Shen, L.; Lehman, L.w.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Anthony Celi, L.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef]
- Faltys, M.; Zimmermann, M.; Lyu, X.; Hüser, M.; Hyland, S.; Rätsch, G.; Merz, T. HiRID, a high time-resolution ICU dataset (version 1.1. 1). PhysioNet 2021. Available online: https://physionet.org/content/hirid/1.1.1/ (accessed on 18 February 2021). [CrossRef]
- Pollard, T.J.; Johnson, A.E.; Raffa, J.D.; Celi, L.A.; Mark, R.G.; Badawi, O. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci. Data 2018, 5, 180178. [Google Scholar] [CrossRef]
- Wagner, D.P.; Draper, E.A. Acute physiology and chronic health evaluation (APACHE II) and Medicare reimbursement. Health Care Financ. Rev. 1984, 1984, 91. [Google Scholar]
- Silva, I.; Moody, G.; Scott, D.J.; Celi, L.A.; Mark, R.G. Predicting in-hospital mortality of icu patients: The physionet/computing in cardiology challenge 2012. In Proceedings of the IEEE 2012 Computing in Cardiology, Kraków, Poland, 9–12 September 2012; pp. 245–248. [Google Scholar]
- Orlandic, L.; Teijeiro, T.; Atienza, D. A Semi-Supervised Algorithm for Improving the Consistency of Crowdsourced Datasets: The COVID-19 Case Study on Respiratory Disorder Classification. arXiv 2022, arXiv:2209.04360. [Google Scholar]
- Müller, M. Dynamic time warping. In Information Retrieval for Music and Motion; Springer: Berlin/Heidelberg, Germany, 2007; pp. 69–84. [Google Scholar]
- Tonekaboni, S.; Eytan, D.; Goldenberg, A. Unsupervised representation learning for time series with temporal neighborhood coding. arXiv 2021, arXiv:2106.00750. [Google Scholar]
- van den Oord, A.; Li, Y.; Vinyals, O. Representation learning with contrastive predictive coding. arXiv 2018, arXiv:1807.03748. [Google Scholar]
- Khaertdinov, B.; Ghaleb, E.; Asteriadis, S. Contrastive self-supervised learning for sensor-based human activity recognition. In Proceedings of the 2021 IEEE International Joint Conference on Biometrics (IJCB), Virtual, 4–7 August 2021; pp. 1–8. [Google Scholar]
- Gutmann, M.; Hyvärinen, A. Noise-contrastive estimation: A new estimation principle for unnormalized statistical models. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics. JMLR Workshop and Conference Proceedings, Sardinia, Italy, 13–15 May 2010; pp. 297–304. [Google Scholar]
- Dong, X.; Shen, J. Triplet loss in siamese network for object tracking. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 459–474. [Google Scholar]
- O’reilly, C.; Gosselin, N.; Carrier, J.; Nielsen, T. Montreal Archive of Sleep Studies: An open-access resource for instrument benchmarking and exploratory research. J. Sleep Res. 2014, 23, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Khalighi, S.; Sousa, T.; Santos, J.M.; Nunes, U. ISRUC-Sleep: A comprehensive public dataset for sleep researchers. Comput. Methods Programs Biomed. 2016, 124, 180–192. [Google Scholar] [CrossRef]
- Malekzadeh, M.; Clegg, R.G.; Cavallaro, A.; Haddadi, H. Mobile sensor data anonymization. In Proceedings of the International Conference on Internet of Things Design and Implementation, Montreal, QC, Canada, 15–18 April 2019; pp. 49–58. [Google Scholar]
- Stisen, A.; Blunck, H.; Bhattacharya, S.; Prentow, T.S.; Kjærgaard, M.B.; Dey, A.; Sonne, T.; Jensen, M.M. Smart devices are different: Assessing and mitigatingmobile sensing heterogeneities for activity recognition. In Proceedings of the 13th ACM Conference on Embedded Networked Sensor Systems, Seoul, Republic of Korea, 1–4 November 2015; pp. 127–140. [Google Scholar]
- Vavoulas, G.; Chatzaki, C.; Malliotakis, T.; Pediaditis, M.; Tsiknakis, M. The mobiact dataset: Recognition of activities of daily living using smartphones. In Proceedings of the International Conference on Information and Communication Technologies for Ageing Well and e-Health, Rome, Italy, 21–22 April 2016; SciTePress: Setúbal, Portugal, 2016; Volume 2, pp. 143–151. [Google Scholar]
- Anguita, D.; Ghio, A.; Oneto, L.; Parra Perez, X.; Reyes Ortiz, J.L. A public domain dataset for human activity recognition using smartphones. In Proceedings of the 21th International European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning, Bruges, Belgium, 24–26 April 2013; pp. 437–442. [Google Scholar]
- Walch, O. Motion and heart rate from a wrist-worn wearable and labeled sleep from polysomnography. PhysioNet 2019, 101. Available online: https://physionet.org/content/sleep-accel/1.0.0/ (accessed on 8 October 2019).
- Westerhuis, M.E.; Visser, G.H.; Moons, K.G.; Van Beek, E.; Benders, M.J.; Bijvoet, S.M.; Van Dessel, H.J.; Drogtrop, A.P.; Van Geijn, H.P.; Graziosi, G.C.; et al. Cardiotocography plus ST analysis of fetal electrocardiogram compared with cardiotocography only for intrapartum monitoring: A randomized controlled trial. Obstet. Gynecol. 2010, 115, 1173–1180. [Google Scholar] [CrossRef]
- Muguli, A.; Pinto, L.; Sharma, N.; Krishnan, P.; Ghosh, P.K.; Kumar, R.; Bhat, S.; Chetupalli, S.R.; Ganapathy, S.; Ramoji, S.; et al. DiCOVA Challenge: Dataset, task, and baseline system for COVID-19 diagnosis using acoustics. arXiv 2021, arXiv:2103.09148. [Google Scholar]
- Ribeiro, A.L.P.; Paixao, G.M.; Gomes, P.R.; Ribeiro, M.H.; Ribeiro, A.H.; Canazart, J.A.; Oliveira, D.M.; Ferreira, M.P.; Lima, E.M.; de Moraes, J.L.; et al. Tele-electrocardiography and bigdata: The CODE (Clinical Outcomes in Digital Electrocardiography) study. J. Electrocardiol. 2019, 57, S75–S78. [Google Scholar] [CrossRef]
- Ribeiro, A.H.; Ribeiro, M.H.; Paixão, G.M.; Oliveira, D.M.; Gomes, P.R.; Canazart, J.A.; Ferreira, M.P.; Andersson, C.R.; Macfarlane, P.W.; Meira, W., Jr.; et al. Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat. Commun. 2020, 11, 1760. [Google Scholar] [CrossRef] [PubMed]
- Alday, E.A.P.; Gu, A.; Shah, A.J.; Robichaux, C.; Wong, A.K.I.; Liu, C.; Liu, F.; Rad, A.B.; Elola, A.; Seyedi, S.; et al. Classification of 12-lead ecgs: The physionet/computing in cardiology challenge 2020. Physiol. Meas. 2020, 41, 124003. [Google Scholar] [CrossRef] [PubMed]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Clifford, G.D.; Liu, C.; Moody, B.; Li-wei, H.L.; Silva, I.; Li, Q.; Johnson, A.; Mark, R.G. AF classification from a short single lead ECG recording: The PhysioNet/computing in cardiology challenge 2017. In Proceedings of the IEEE 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar]
- Liu, F.; Liu, C.; Zhao, L.; Zhang, X.; Wu, X.; Xu, X.; Liu, Y.; Ma, C.; Wei, S.; He, Z.; et al. An open access database for evaluating the algorithms of electrocardiogram rhythm and morphology abnormality detection. J. Med Imaging Health Inform. 2018, 8, 1368–1373. [Google Scholar] [CrossRef]
- Bousseljot, R.; Kreiseler, D.; Schnabel, A. Nutzung der EKG-Signaldatenbank CARDIODAT der PTB über Das Internet. Biomedizinische Technik. Biomedizinische Technik, Band 40, Ergänzungsband 1 (1995) S 317. Available online: https://archive.physionet.org/physiobank/database/ptbdb/ (accessed on 8 October 2019).
- Zheng, J.; Zhang, J.; Danioko, S.; Yao, H.; Guo, H.; Rakovski, C. A 12-lead electrocardiogram database for arrhythmia research covering more than 10,000 patients. Sci. Data 2020, 7, 48. [Google Scholar] [CrossRef]
- Hannun, A.Y.; Rajpurkar, P.; Haghpanahi, M.; Tison, G.H.; Bourn, C.; Turakhia, M.P.; Ng, A.Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019, 25, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Wagner, P.; Strodthoff, N.; Bousseljot, R.-D.; Kreiseler, D.; Lunze, F.I.; Samek, W.; Schaeffter, T. A. PTB-XL, a large publicly available electrocardiography dataset. Sci. Data 2020, 7, 154. [Google Scholar] [CrossRef]
- Greenwald, S.D.; Patil, R.S.; Mark, R.G. Improved Detection and Classification of Arrhythmias in Noise-Corrupted Electrocardiograms Using Contextual Information. In Proceedings of the [1990] Proceedings Computers in Cardiology, Chicago, IL, USA, 23–26 September 1990. [Google Scholar]
- Stuldreher, I.V.; Thammasan, N.; van Erp, J.B.; Brouwer, A.M. Physiological synchrony in EEG, electrodermal activity and heart rate reflects shared selective auditory attention. J. Neural Eng. 2020, 17, 046028. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Cui, L.; Mueller, R.; Tao, S.; Kim, M.; Rueschman, M.; Mariani, S.; Mobley, D.; Redline, S. The National Sleep Research Resource: Towards a sleep data commons. J. Am. Med Inform. Assoc. 2018, 25, 1351–1358. [Google Scholar] [CrossRef]
- Quan, S.F.; Howard, B.V.; Iber, C.; Kiley, J.P.; Nieto, F.J.; O’Connor, G.T.; Rapoport, D.M.; Redline, S.; Robbins, J.; Samet, J.M.; et al. The sleep heart health study: Design, rationale, and methods. Sleep 1997, 20, 1077–1085. [Google Scholar]
- Miranda-Correa, J.A.; Abadi, M.K.; Sebe, N.; Patras, I. Amigos: A dataset for affect, personality and mood research on individuals and groups. IEEE Trans. Affect. Comput. 2018, 12, 479–493. [Google Scholar] [CrossRef]
- Babayan, A.; Erbey, M.; Kumral, D.; Reinelt, J.D.; Reiter, A.M.; Röbbig, J.; Schaare, H.L.; Uhlig, M.; Anwander, A.; Bazin, P.L.; et al. A mind-brain-body dataset of MRI, EEG, cognition, emotion, and peripheral physiology in young and old adults. Sci. Data 2019, 6, 180308. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.; Reiss, A.; Duerichen, R.; Marberger, C.; Van Laerhoven, K. Introducing wesad, a multimodal dataset for wearable stress and affect detection. In Proceedings of the 20th ACM International Conference on Multimodal Interaction, Boulder, CO, USA, 16–20 October 2018; pp. 400–408. [Google Scholar]
- Koldijk, S.; Sappelli, M.; Verberne, S.; Neerincx, M.A.; Kraaij, W. The swell knowledge work dataset for stress and user modeling research. In Proceedings of the 16th International Conference on Multimodal Interaction, Istanbul, Turkey, 12–16 November 2014; pp. 291–298. [Google Scholar]
- O’Connor, L.; Brage, S.; Griffin, S.J.; Wareham, N.J.; Forouhi, N.G. The cross-sectional association between snacking behaviour and measures of adiposity: The Fenland Study, UK. Br. J. Nutr. 2015, 114, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.L.; Lu, B.L. Investigating Critical Frequency Bands and Channels for EEG-based Emotion Recognition with Deep Neural Networks. IEEE Trans. Auton. Ment. Dev. 2015, 7, 162–175. [Google Scholar] [CrossRef]
- Duan, R.N.; Zhu, J.Y.; Lu, B.L. Differential entropy feature for EEG-based emotion classification. In Proceedings of the 6th International IEEE/EMBS Conference on Neural Engineering (NER), San Diego, CA, USA, 6–8 November 2013; pp. 81–84. [Google Scholar]
- Obeid, I.; Picone, J. The temple university hospital EEG data corpus. Front. Neurosci. 2016, 10, 196. [Google Scholar] [CrossRef]
- Schalk, G.; McFarland, D.J.; Hinterberger, T.; Birbaumer, N.; Wolpaw, J.R. BCI2000: A general-purpose brain-computer interface (BCI) system. IEEE Trans. Biomed. Eng. 2004, 51, 1034–1043. [Google Scholar] [CrossRef]
- Brunner, C.; Leeb, R.; Müller-Putz, G.; Schlögl, A.; Pfurtscheller, G. BCI Competition 2008–Graz data set A. In Institute for Knowledge Discovery (Laboratory of Brain-Computer Interfaces); Graz University of Technology: Styria, Austria, 2008; Volume 16, pp. 1–6. [Google Scholar]
- Kemp, B.; Zwinderman, A.H.; Tuk, B.; Kamphuisen, H.A.; Oberye, J.J. Analysis of a sleep-dependent neuronal feedback loop: The slow-wave microcontinuity of the EEG. IEEE Trans. Biomed. Eng. 2000, 47, 1185–1194. [Google Scholar] [CrossRef]
- Goldberger, A.; Amaral, L.; Glass, L.; Hausdorff, J.; Ivanov, P.C.; Mark, R.; Mietus, J.; Moody, G.; Peng, C.; Stanley, H. PhysioBank, PhysioToolkit, and Physionet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef]
- Ma, X.; Qiu, S.; He, H. Multi-channel EEG recording during motor imagery of different joints from the same limb. Sci. Data 2020, 7, 191. [Google Scholar] [CrossRef]
- Ihle, M.; Feldwisch-Drentrup, H.; Teixeira, C.A.; Witon, A.; Schelter, B.; Timmer, J.; Schulze-Bonhage, A. EPILEPSIAE—A European epilepsy database. Comput. Methods Programs Biomed. 2012, 106, 127–138. [Google Scholar] [CrossRef]
- Temko, A.; Sarkar, A.; Lightbody, G. Detection of seizures in intracranial EEG: UPenn and Mayo Clinic’s Seizure detection challenge. In Proceedings of the EMBC, Milan, Italy, 25–29 August 2015; pp. 6582–6585. [Google Scholar]
- Guillot, A.; Sauvet, F.; During, E.H.; Thorey, V. Dreem open datasets: Multi-scored sleep datasets to compare human and automated sleep staging. IEEE Trans. Neural Syst. Rehabil. Eng. 2020, 28, 1955–1965. [Google Scholar] [CrossRef] [PubMed]
- Katsigiannis, S.; Ramzan, N. DREAMER: A database for emotion recognition through EEG and ECG signals from wireless low-cost off-the-shelf devices. IEEE J. Biomed. Health Inform. 2017, 22, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Ghassemi, M.M.; Moody, B.E.; Lehman, L.W.H.; Song, C.; Li, Q.; Sun, H.; Mark, R.G.; Westover, M.B.; Clifford, G.D. You snooze, you win: The physionet/computing in cardiology challenge 2018. In Proceedings of the IEEE 2018 Computing in Cardiology Conference (CinC), Maastricht, The Netherlands, 23–26 September 2018; Volume 45, pp. 1–4. [Google Scholar]
- Hyland, S.L.; Faltys, M.; Hüser, M.; Lyu, X.; Gumbsch, T.; Esteban, C.; Bock, C.; Horn, M.; Moor, M.; Rieck, B.; et al. Machine learning for early prediction of circulatory failure in the intensive care unit. arXiv 2019, arXiv:1904.07990. [Google Scholar] [CrossRef] [PubMed]
- Reyna, M.A.; Kiarashi, Y.; Elola, A.; Oliveira, J.; Renna, F.; Gu, A.; Perez-Alday, E.A.; Sadr, N.; Sharma, A.; Mattos, S.; et al. Heart murmur detection from phonocardiogram recordings: The george b. moody physionet challenge 2022. medRxiv 2022. [Google Scholar] [CrossRef]
- Tan, C.; Zhang, L.; Wu, H.t. A novel Blaschke unwinding adaptive-Fourier-decomposition-based signal compression algorithm with application on ECG signals. IEEE J. Biomed. Health Inform. 2018, 23, 672–682. [Google Scholar] [CrossRef]
- Rocha, B.; Filos, D.; Mendes, L.; Vogiatzis, I.; Perantoni, E.; Kaimakamis, E.; Natsiavas, P.; Oliveira, A.; Jácome, C.; Marques, A.; et al. A respiratory sound database for the development of automated classification. In Proceedings of the International Conference on Biomedical and Health Informatics; Springer: Berlin/Heidelberg, Germany, 2018; pp. 33–37. [Google Scholar]
- Panayotov, V.; Chen, G.; Povey, D.; Khudanpur, S. Librispeech: An asr corpus based on public domain audio books. In Proceedings of the 2015 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brisbane, QLD, Australia, 19–24 April 2015; pp. 5206–5210. [Google Scholar]
- Zhao, R.; Yan, R.; Chen, Z.; Mao, K.; Wang, P.; Gao, R.X. Deep learning and its applications to machine health monitoring. Mech. Syst. Signal Process. 2019, 115, 213–237. [Google Scholar] [CrossRef]
- Yi, X.; Stokes, D.; Yan, Y.; Liao, C. CUDAMicroBench: Microbenchmarks to Assist CUDA Performance Programming. In Proceedings of the IPDPSW, Portland, OR, USA, 17–21 June 2021; pp. 397–406. [Google Scholar]
- He, K.; Fan, H.; Wu, Y.; Xie, S.; Girshick, R. Momentum contrast for unsupervised visual representation learning. In Proceedings of the IEEE/CVF CVPR, Virtual, 14–19 June 2020; pp. 9729–9738. [Google Scholar]




| Database | Query | Articles Returned |
|---|---|---|
| IEEE | (((self-supervised) OR (Contrastive)) AND ((“medical time series”) OR (“physiological signal”) OR (“biomedical signal”) OR (“medical signal”) OR “biosignal”)) | 189 |
| ACM | 529 | |
| Scopus | 60 | |
| Google Scholar | 1285 | |
| MEDLINE (PubMed) | (((self-supervised) OR (“Contrastive learning”)) AND ((medical time series) OR (physiological signal) OR (biomedical signal) OR (medical signal))) | 39 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Alavi, A.; Li, M.; Zhang, X. Self-Supervised Contrastive Learning for Medical Time Series: A Systematic Review. Sensors 2023, 23, 4221. https://doi.org/10.3390/s23094221
Liu Z, Alavi A, Li M, Zhang X. Self-Supervised Contrastive Learning for Medical Time Series: A Systematic Review. Sensors. 2023; 23(9):4221. https://doi.org/10.3390/s23094221
Chicago/Turabian StyleLiu, Ziyu, Azadeh Alavi, Minyi Li, and Xiang Zhang. 2023. "Self-Supervised Contrastive Learning for Medical Time Series: A Systematic Review" Sensors 23, no. 9: 4221. https://doi.org/10.3390/s23094221
APA StyleLiu, Z., Alavi, A., Li, M., & Zhang, X. (2023). Self-Supervised Contrastive Learning for Medical Time Series: A Systematic Review. Sensors, 23(9), 4221. https://doi.org/10.3390/s23094221






