A Deep Learning Framework for Processing and Classification of Hyperspectral Rice Seed Images Grown under High Day and Night Temperatures
Abstract
1. Introduction
- (a)
- Classification of HSIs of rice seeds grown under different exposure durations to different high day and/or night temperature treatments using DL architectures;
- (b)
- A DL framework for a comprehensive analysis of hyperspectral images of seeds. We present results from rice seeds grown under High Night Temperature (HNT), High Day and High Night Temperature (HDNT) stressors, and control environments. Our framework includes options for calibration, preprocessing, and segmentation for HSI seed image extraction from a panel image of seeds, spectral analysis, spatial–spectral feature extraction, as well as classification using two different DL neural network architectures: 3D-CNN and Deep Neural Network (DNN);
- (c)
- A software application of the DL seed image processing framework is the first of its kind for processing crop seed HSIs.
2. Materials and Methods
2.1. Hyperspectral Rice Seed Datasets
2.2. Calibration and Segmentation of Rice Seed HSI
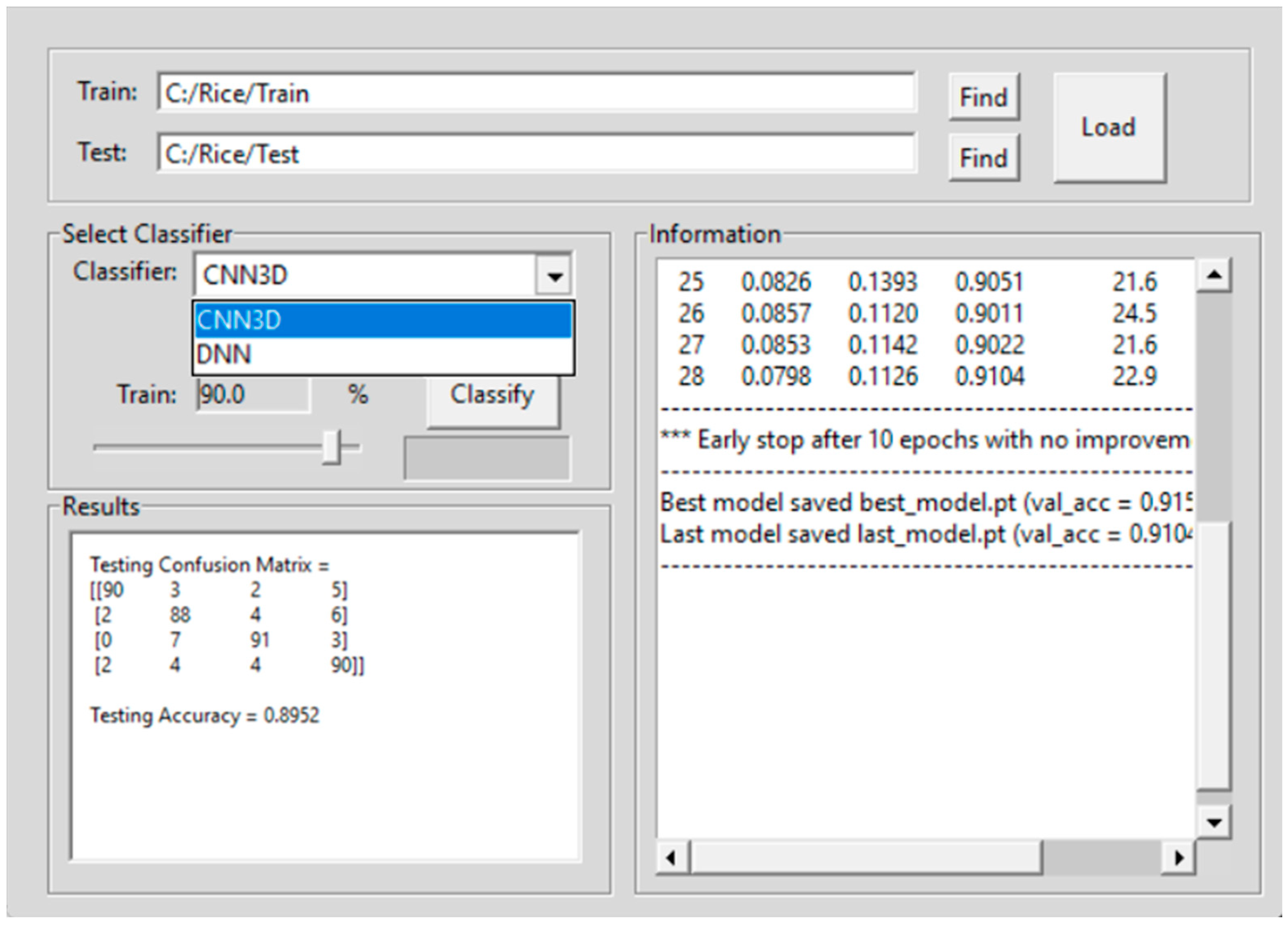
2.3. Hyperspectral Seed Graphical User Interfaces
2.4. 3D-Convolutional Neural Network
2.5. 3D-CNN Training and Validations
2.6. Deep Neural Network
2.7. DNN Training and Validation
3. Results
4. Discussion
Comparison with State-of-the-Art Rice Seed HSI Classification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Li, P.; Chen, Y.H.; Lu, J.; Zhang, C.Q.; Liu, Q.Q.; Li, Q.F. Genes and Their Molecular Functions Determining Seed Structure, Components, and Quality of Rice. Rice 2022, 15, 18. [Google Scholar] [CrossRef] [PubMed]
- Dhatt, B.K.; Abshire, N.; Paul, P.; Hasanthika, K.; Sandhu, J.; Zhang, Q.; Obata, T.; Walia, H. Metabolic Dynamics of Developing Rice Seeds Under High Night-Time Temperature Stress. Front. Plant Sci. 2019, 10, 1443. [Google Scholar] [CrossRef] [PubMed]
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Comput. Sci. 2021, 2, 420. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Tan, Q.; Zhang, Y.; Zha, X.; Yang, S.; Yang, R. Research on Maize Seed Classification and Recognition Based on Machine Vision and Deep Learning. Agriculture 2022, 12, 232. [Google Scholar] [CrossRef]
- Ruslan, R.; Khairunniza-Bejo, S.; Jahari, M.; Ibrahim, M.F. Weedy Rice Classification Using Image Processing and a Machine Learning Approach. Agriculture 2022, 12, 645. [Google Scholar] [CrossRef]
- Elmasry, G.; Mandour, N.; Al-Rejaie, S.; Belin, E.; Rousseau, D. Recent applications of multispectral imaging in seed phenotyping and quality monitoring—An overview. Sensors 2019, 19, 1090. [Google Scholar] [CrossRef] [PubMed]
- Fabiyi, S.D.; Vu, H.; Tachtatzis, C.; Murray, P.; Harle, D.; Dao, T.K.; Andonovic, I.; Ren, J.; Marshall, S. Varietal Classification of Rice Seeds Using RGB and Hyperspectral Images. IEEE Access 2020, 8, 22493–22505. [Google Scholar] [CrossRef]
- Qiu, Z.; Chen, J.; Zhao, Y.; Zhu, S.; He, Y.; Zhang, C. Variety identification of single rice seed using hyperspectral imaging combined with convolutional neural network. Appl. Sci. 2018, 8, 212. [Google Scholar] [CrossRef]
- Onmankhong, J.; Ma, T.; Inagaki, T.; Sirisomboon, P.; Tsuchikawa, S. Cognitive spectroscopy for the classification of rice varieties: A comparison of machine learning and deep learning approaches in analysing long-wave near-infrared hyperspectral images of brown and milled samples. Infrared Phys. Technol. 2022, 123, 104100. [Google Scholar] [CrossRef]
- Zhou, S.; Sun, L.; Xing, W.; Feng, G.; Ji, Y.; Yang, J.; Liu, S. Hyperspectral imaging of beet seed germination prediction. Infrared Phys. Technol. 2020, 108, 103363. [Google Scholar] [CrossRef]
- Chatnuntawech, I.; Tantisantisom, K.; Khanchaitit, P.; Boonkoom, T.; Bilgic, B.; Chuangsuwanich, E. Rice Classification Using Spatio-Spectral Deep Convolutional Neural Network. arXiv 2018, arXiv:1805.11491. [Google Scholar]
- Dar, R.A.; Bhat, D.; Assad, A.; Islam, Z.U.; Gulzar, W.; Yaseen, A. Classification of Rice Grain Varieties Using Deep Convolutional Neural Network Architectures. SSRN Electron. J. 2022. [Google Scholar] [CrossRef]
- Wu, N.; Liu, F.; Meng, F.; Li, M.; Zhang, C.; He, Y. Rapid and Accurate Varieties Classification of Different Crop Seeds Under Sample-Limited Condition Based on Hyperspectral Imaging and Deep Transfer Learning. Front. Bioeng. Biotechnol. 2021, 9, 696292. [Google Scholar] [CrossRef]
- Gao, T.; Chandran, A.K.N.; Paul, P.; Walia, H.; Yu, H. HyperSeed: An End-to-End Method to Process Hyperspectral Images of Seeds. Sensors 2021, 21, 8184. [Google Scholar] [CrossRef]
- Polder, G.; Van Der Heijden, G.W.; Keizer, L.P.; Young, I.T. Calibration and Characterisation of Imaging Spectrographs. J. Near Infrared Spectrosc. 2003, 11, 193–210. [Google Scholar] [CrossRef]
- Woods, R.E.; Gonzalez, R.C. Digital Image Processing; Pearson: London, UK, 2019. [Google Scholar]
- Satoto, B.D.; Anamisa, D.R.; Yusuf, M.; Sophan, M.K.; Khairunnisa, S.O.; Irmawati, B. Rice seed classification using machine learning and deep learning. In Proceedings of the 2022 7th International Conference Informatics Computer ICIC 2022, Denpasar, Indonesia, 8–9 December 2022. [Google Scholar] [CrossRef]
- Aukkapinyo, K.; Sawangwong, S.; Pooyoi, P.; Kusakunniran, W. Localization and Classification of Rice-grain Images Using Region Proposals-based Convolutional Neural Network. Int. J. Autom. Comput. 2020, 17, 233–246. [Google Scholar] [CrossRef]
- Panmuang, M.; Rodmorn, C.; Pinitkan, S. Image Processing for Classification of Rice Varieties with Deep Convolutional Neural Networks. In Proceedings of the 16th International Symposium on Artificial Intelligence and Natural Language Language Processing, iSAI-NLP 2021, Ayutthaya, Thailand, 21–23 December 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Tuğrul, B. Classification of Five Different Rice Seeds Grown. Commun. Fac. Sci. Univ. Ank. Ser. A2-A3 Phys. Sci. Eng. 2022, 64, 40–50. [Google Scholar] [CrossRef]
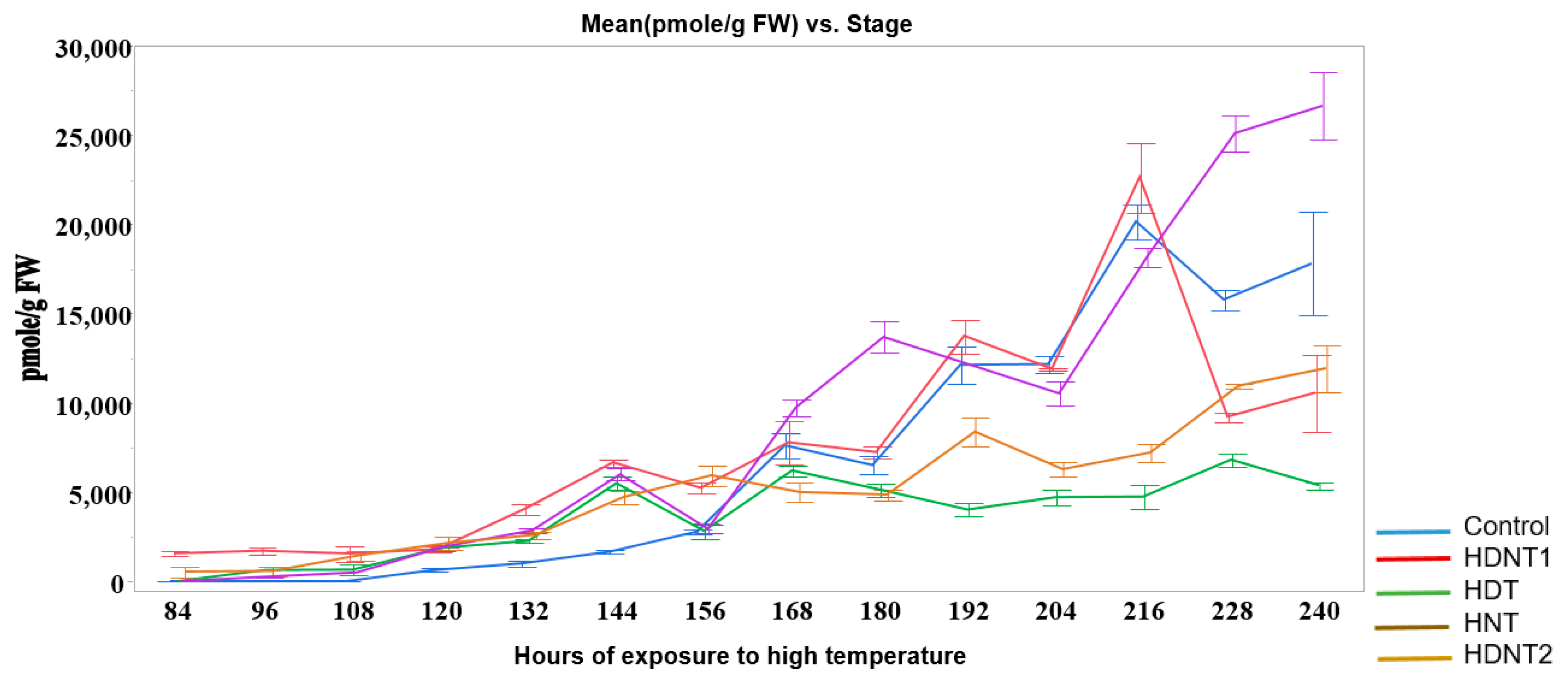
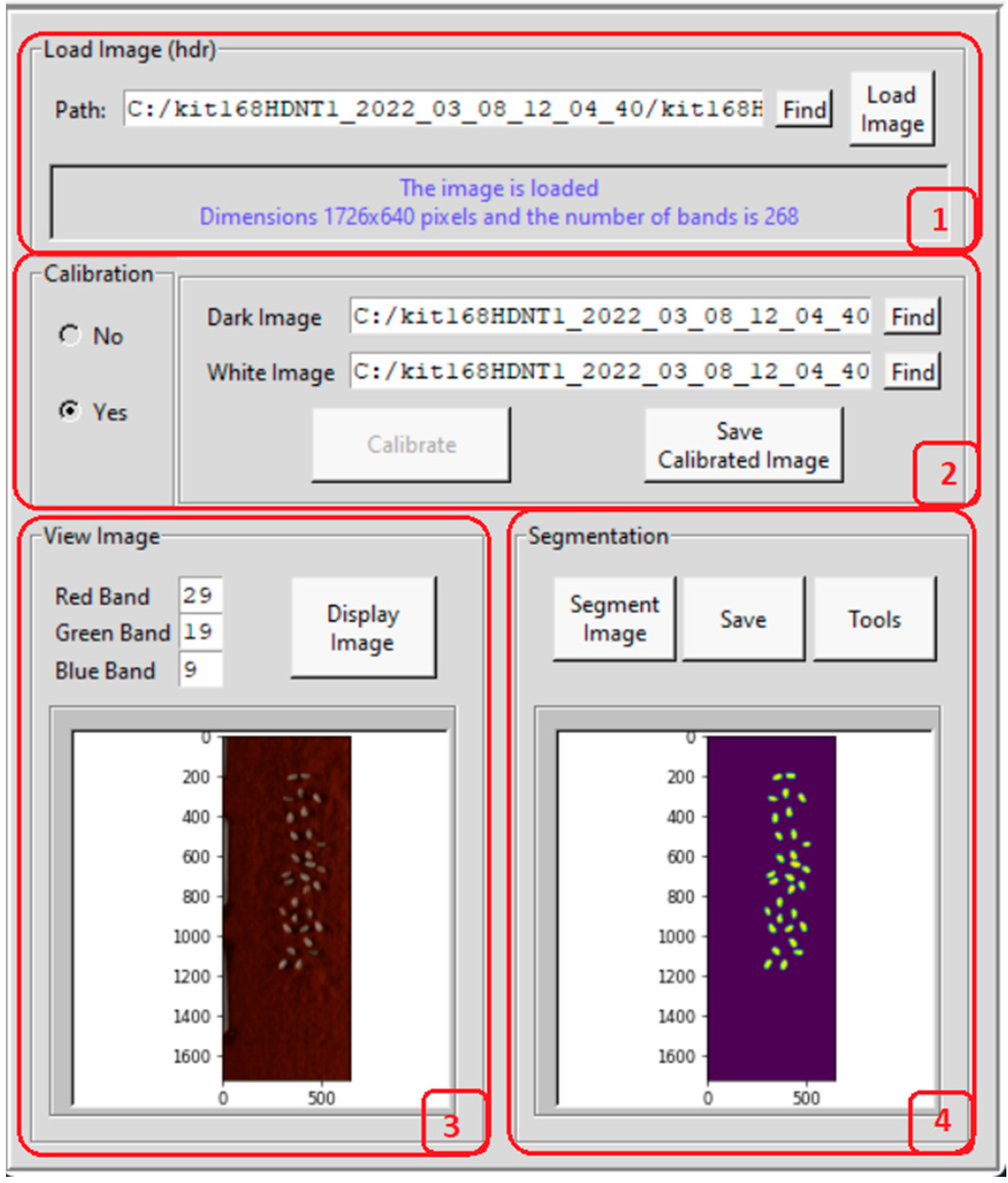
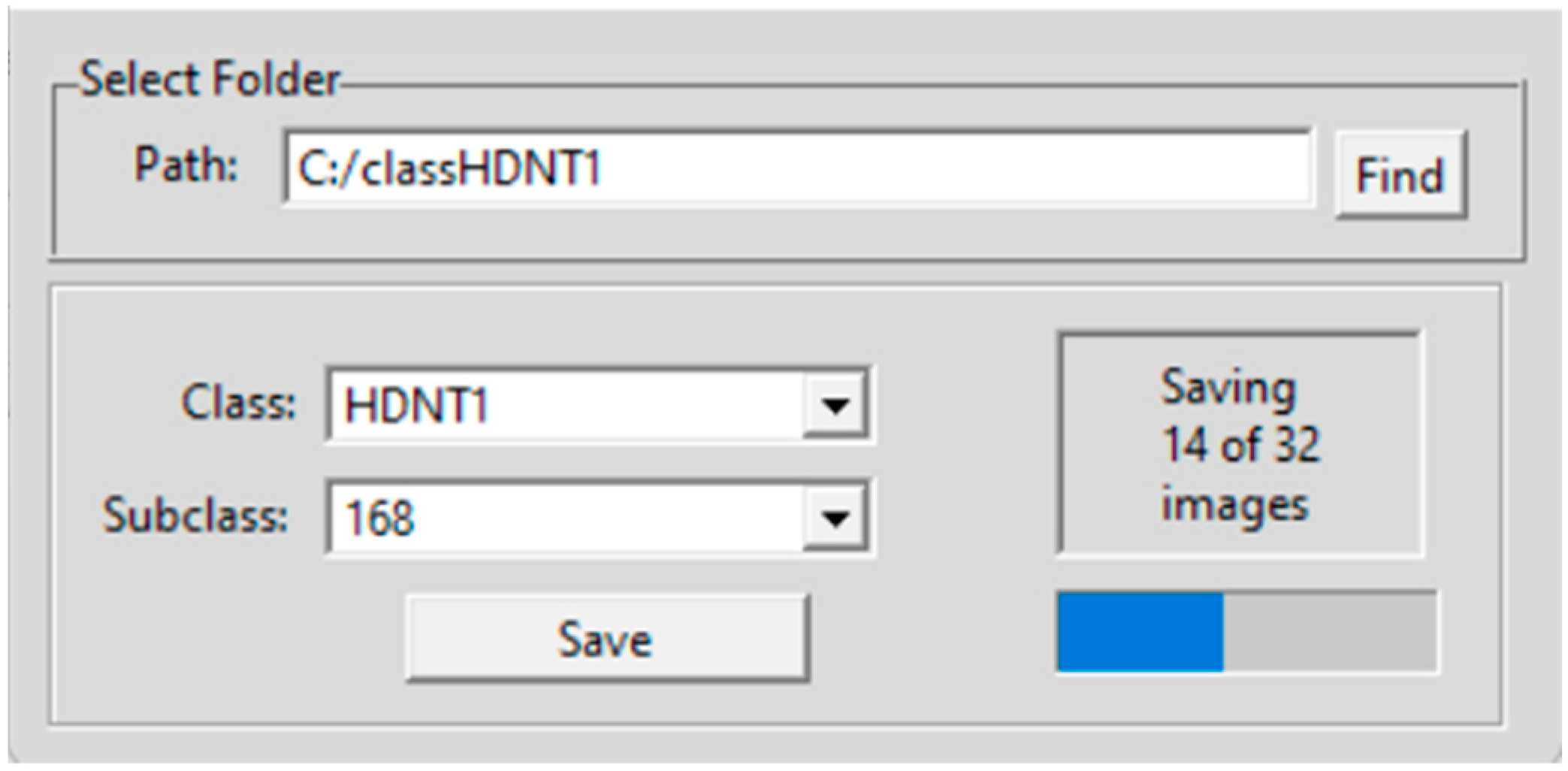
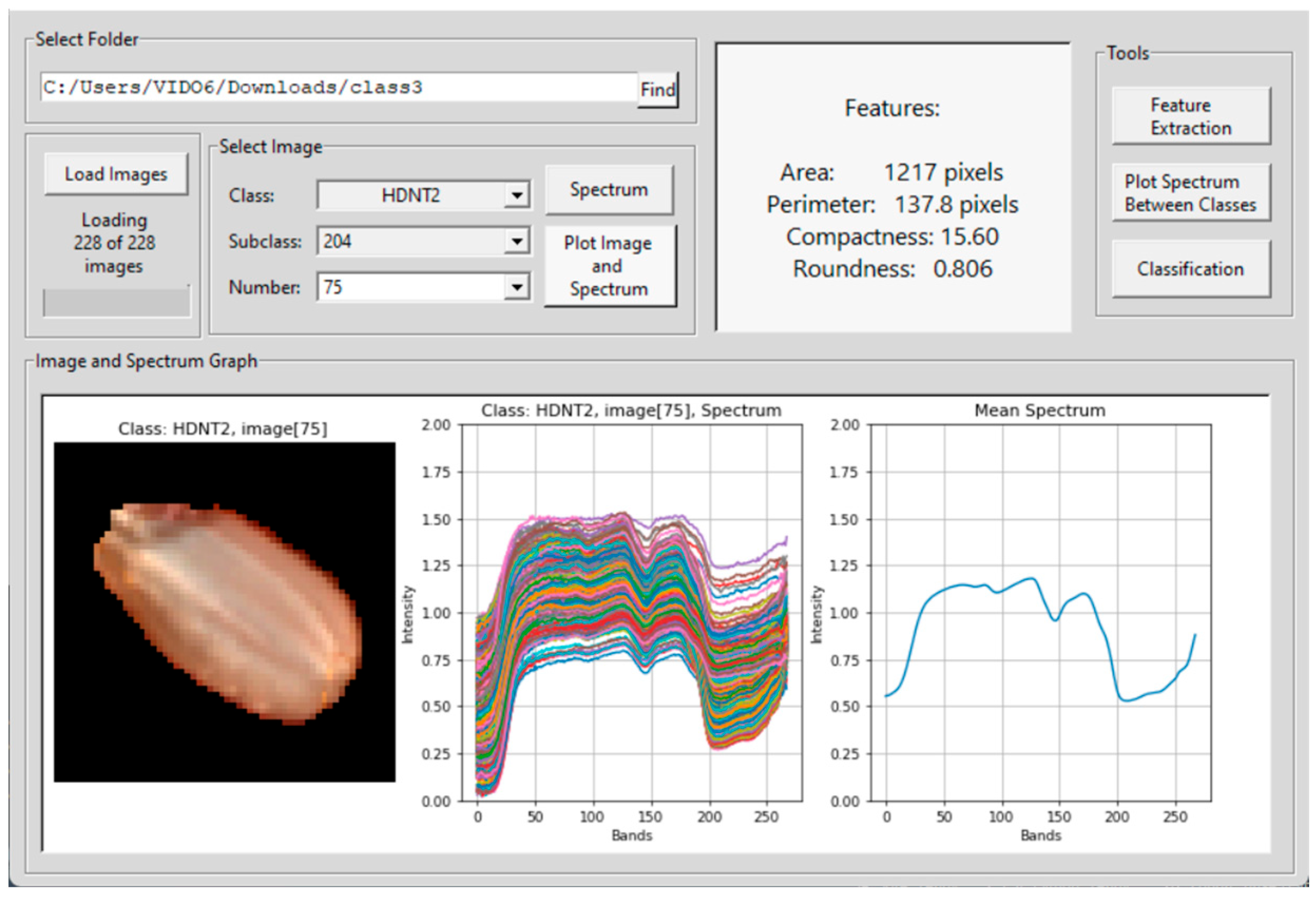


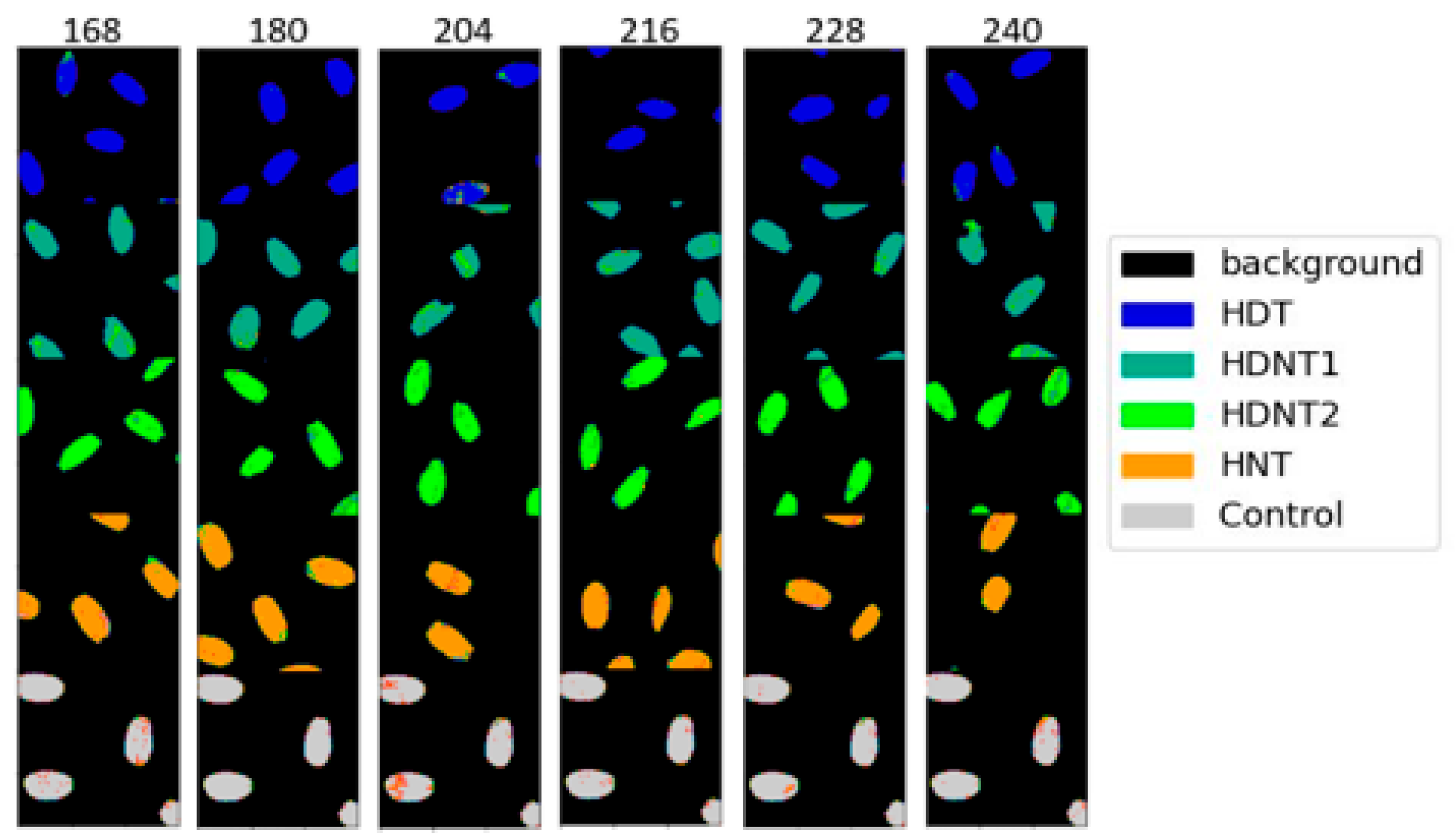

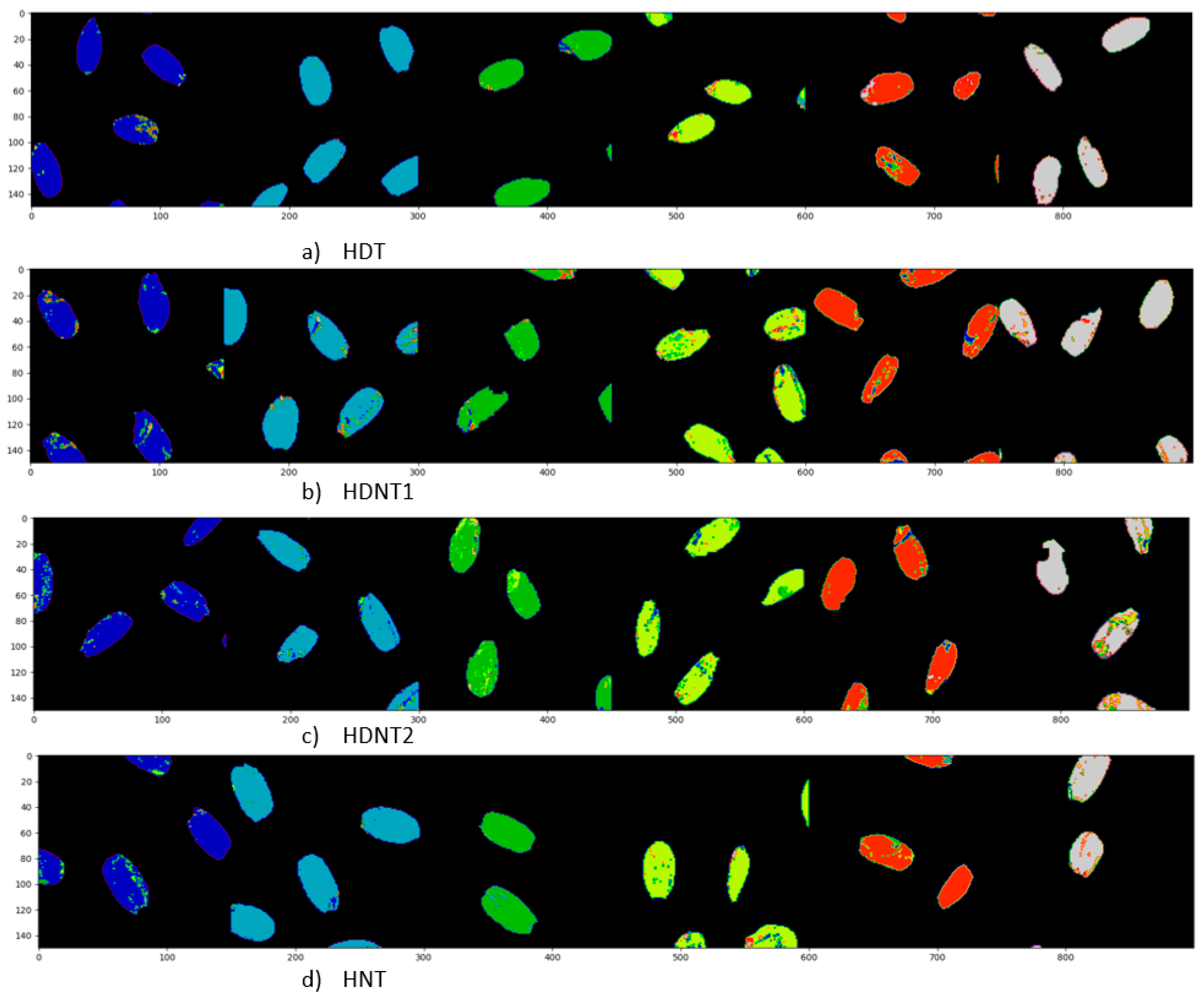


| Treatment Class | Day/Night Temperature °C | Number of Images |
|---|---|---|
| Control | 28/23 | 40 |
| High day/night temperature 1 (HDNT1) | 36/32 | 40 |
| High night temperature (HNT) | 28/28 | 40 |
| High day temperature (HDT) | 36/23 | 40 |
| High day/night temperature (HDNT2) | 36/28 | 40 |
| Performance Metrics | Formula |
|---|---|
| Precision | |
| Recall | |
| F1-Score | |
| Average Accuracy (AA) | |
| Overall Accuracy (OA) | |
| Kappa (K) |
| High Temperature Treatment Classes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Exposure | HDT | HDNT1 | HDNT2 | HNT | Control | AA | OA | K |
| 168 | 0.93 | 0.91 | 0.93 | 0.93 | 0.92 | 0.92 | 0.91 | 0.91 |
| 180 | 0.91 | 0.9 | 0.89 | 0.92 | 0.89 | 0.91 | 0.9 | 0.89 |
| 204 | 0.94 | 0.93 | 0.93 | 0.94 | 0.94 | 0.94 | 0.94 | 0.94 |
| 216 | 0.92 | 0.91 | 0.91 | 0.9 | 0.93 | 0.92 | 0.92 | 0.91 |
| 228 | 0.91 | 0.91 | 0.9 | 0.91 | 0.91 | 0.91 | 0.9 | 0.91 |
| 240 | 0.92 | 0.92 | 0.92 | 0.93 | 0.92 | 0.92 | 0.91 | 0.92 |
| High Temperature Exposure Time (Hours) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Classes | 168 | 180 | 204 | 216 | 228 | 240 | AA | OA | K |
| HDNT1 | 0.89 | 0.88 | 0.88 | 0.89 | 0.89 | 0.89 | 0.89 | 0.89 | 0.88 |
| HDNT2 | 0.91 | 0.9 | 0.92 | 0.9 | 0.89 | 0.9 | 0.9 | 0.89 | 0.88 |
| HDT | 0.91 | 0.92 | 0.92 | 0.9 | 0.91 | 0.9 | 0.91 | 0.91 | 0.9 |
| HNT | 0.91 | 0.92 | 0.92 | 0.9 | 0.91 | 0.91 | 0.91 | 0.9 | 0.89 |
| 168 h | 180 h | 204 h | 216 h | 228 h | 240 h | |
|---|---|---|---|---|---|---|
| Classes | Precision | Precision | Precision | Precision | Precision | Precision |
| HDT | 0.97 | 1 | 0.92 | 1 | 0.99 | 0.98 |
| HDNT1 | 0.92 | 0.96 | 0.85 | 0.96 | 0.97 | 0.86 |
| HDNT2 | 0.95 | 0.9 | 0.93 | 0.91 | 0.96 | 0.86 |
| HNT | 0.93 | 0.96 | 0.95 | 0.96 | 0.92 | 0.96 |
| Control | 0.96 | 1 | 0.94 | 0.98 | 0.98 | 0.96 |
| Macro average | 0.95 | 0.96 | 0.92 | 0.96 | 0.96 | 0.92 |
| Weighted average | 0.95 | 0.97 | 0.92 | 0.96 | 0.97 | 0.92 |
| Classes | recall | recall | recall | recall | recall | recall |
| HDT | 0.97 | 1 | 0.98 | 0.96 | 1 | 1 |
| HDNT1 | 0.97 | 0.91 | 0.86 | 0.96 | 0.95 | 0.86 |
| HDNT2 | 0.88 | 0.95 | 0.89 | 0.95 | 0.96 | 0.84 |
| HNT | 0.95 | 0.99 | 0.88 | 0.95 | 0.96 | 0.9 |
| Control | 0.96 | 0.99 | 0.96 | 0.99 | 0.97 | 0.99 |
| Macro average | 0.94 | 0.97 | 0.92 | 0.96 | 0.97 | 0.92 |
| Weighted average | 0.95 | 0.97 | 0.92 | 0.96 | 0.97 | 0.92 |
| Classes | f1-score | f1-score | f1-score | f1-score | f1-score | f1-score |
| HDT | 0.97 | 1 | 0.95 | 0.98 | 1 | 0.99 |
| HDNT1 | 0.94 | 0.94 | 0.85 | 0.96 | 0.96 | 0.86 |
| HDNT2 | 0.91 | 0.92 | 0.91 | 0.93 | 0.96 | 0.85 |
| HNT | 0.94 | 0.97 | 0.92 | 0.96 | 0.94 | 0.93 |
| Control | 0.96 | 0.99 | 0.95 | 0.98 | 0.97 | 0.97 |
| Accuracy | 0.95 | 0.97 | 0.92 | 0.96 | 0.97 | 0.92 |
| Macro average | 0.94 | 0.96 | 0.91 | 0.96 | 0.97 | 0.92 |
| Weighted average | 0.95 | 0.97 | 0.92 | 0.96 | 0.97 | 0.92 |
| HDT | HDNT1 | HDNT2 | HNT | |
|---|---|---|---|---|
| Classes | Precision | Precision | Precision | Precision |
| 168 | 0.95 | 0.89 | 0.91 | 0.91 |
| 180 | 0.99 | 0.91 | 0.93 | 0.98 |
| 204 | 0.96 | 0.86 | 0.81 | 0.94 |
| 216 | 0.86 | 0.83 | 0.86 | 0.89 |
| 228 | 0.86 | 0.9 | 0.9 | 0.92 |
| 240 | 0.98 | 0.94 | 0.78 | 0.91 |
| Macro average | 0.94 | 0.89 | 0.87 | 0.93 |
| Weighted average | 0.95 | 0.89 | 0.87 | 0.93 |
| Classes | recall | recall | recall | recall |
| 168 | 0.96 | 0.83 | 0.83 | 0.92 |
| 180 | 0.97 | 0.98 | 0.92 | 0.96 |
| 204 | 0.96 | 0.87 | 0.88 | 0.92 |
| 216 | 0.99 | 0.92 | 0.76 | 0.89 |
| 228 | 0.9 | 0.82 | 0.9 | 0.96 |
| 240 | 0.91 | 0.89 | 0.93 | 0.91 |
| Macro average | 0.95 | 0.89 | 0.87 | 0.93 |
| Weighted average | 0.95 | 0.89 | 0.87 | 0.93 |
| Classes | f1-score | f1-score | f1-score | f1-score |
| 168 | 0.95 | 0.86 | 0.87 | 0.91 |
| 180 | 0.98 | 0.94 | 0.92 | 0.97 |
| 204 | 0.96 | 0.87 | 0.84 | 0.93 |
| 216 | 0.92 | 0.87 | 0.81 | 0.89 |
| 228 | 0.88 | 0.86 | 0.9 | 0.94 |
| Accuracy | 0.95 | 0.89 | 0.87 | 0.93 |
| Macro average | 0.94 | 0.89 | 0.87 | 0.93 |
| Weighted average | 0.95 | 0.89 | 0.87 | 0.93 |
| Year | Authors | Algorithm | Number Classes | Overall Accuracy |
|---|---|---|---|---|
| 2021 | T. Gao et al. [14] | Rice seed HSI classification using 3D-CNN (high temperature) | 2 | 97.5% |
| 2021 | T. Gao et al. [14] | Rice pixel based HSI classification (high temperature) | 2 | 94.21% |
| 2019 | Z. Qui et al. [8] | Regular rice seed HSI image classification using CNN | 4 | 87% |
| 2018 | I.Hatnuntawech et al. [11] | Regular rice seed HSI classification using ResNet-B | 6 | 91.09% |
| Proposed method | High temperature grown rice seed HSI using 3D-CNN | 6 | 91.33% | |
| Proposed method | High temperature grown rice seed HSI using DNN | 6 | 94.83% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-Martínez, V.; Orozco-Sandoval, J.; Manian, V.; Dhatt, B.K.; Walia, H. A Deep Learning Framework for Processing and Classification of Hyperspectral Rice Seed Images Grown under High Day and Night Temperatures. Sensors 2023, 23, 4370. https://doi.org/10.3390/s23094370
Díaz-Martínez V, Orozco-Sandoval J, Manian V, Dhatt BK, Walia H. A Deep Learning Framework for Processing and Classification of Hyperspectral Rice Seed Images Grown under High Day and Night Temperatures. Sensors. 2023; 23(9):4370. https://doi.org/10.3390/s23094370
Chicago/Turabian StyleDíaz-Martínez, Víctor, Jairo Orozco-Sandoval, Vidya Manian, Balpreet K. Dhatt, and Harkamal Walia. 2023. "A Deep Learning Framework for Processing and Classification of Hyperspectral Rice Seed Images Grown under High Day and Night Temperatures" Sensors 23, no. 9: 4370. https://doi.org/10.3390/s23094370
APA StyleDíaz-Martínez, V., Orozco-Sandoval, J., Manian, V., Dhatt, B. K., & Walia, H. (2023). A Deep Learning Framework for Processing and Classification of Hyperspectral Rice Seed Images Grown under High Day and Night Temperatures. Sensors, 23(9), 4370. https://doi.org/10.3390/s23094370







