Enhancing IoT Healthcare with Federated Learning and Variational Autoencoder
Abstract
1. Introduction
2. Related Work
2.1. Grouping in Federated Learning
2.2. Federated Learning under Heterogeneous Environment
2.3. Contributions
- A performance-efficient VAE-based method is proposed that extracts and learns patients’ features for forming groups and accounts for data heterogeneity.
- The global model is updated while reducing the prediction loss and enhancing classification accuracy. It is evaluated through an extensive set of simulations using a federated learning simulator, with varying scenarios of data heterogeneity, and shows that it outperforms conventional methods.
- The proposed approach takes into account the characteristics of patient data and performs aggregation in a way that achieves the highest accuracy and the lowest loss, making it robust compared to conventional methods. This is accomplished through accurately aggregating local models trained by medical centers and hospitals, considering various parameters such as their latent spaces, data volume, and data variance.
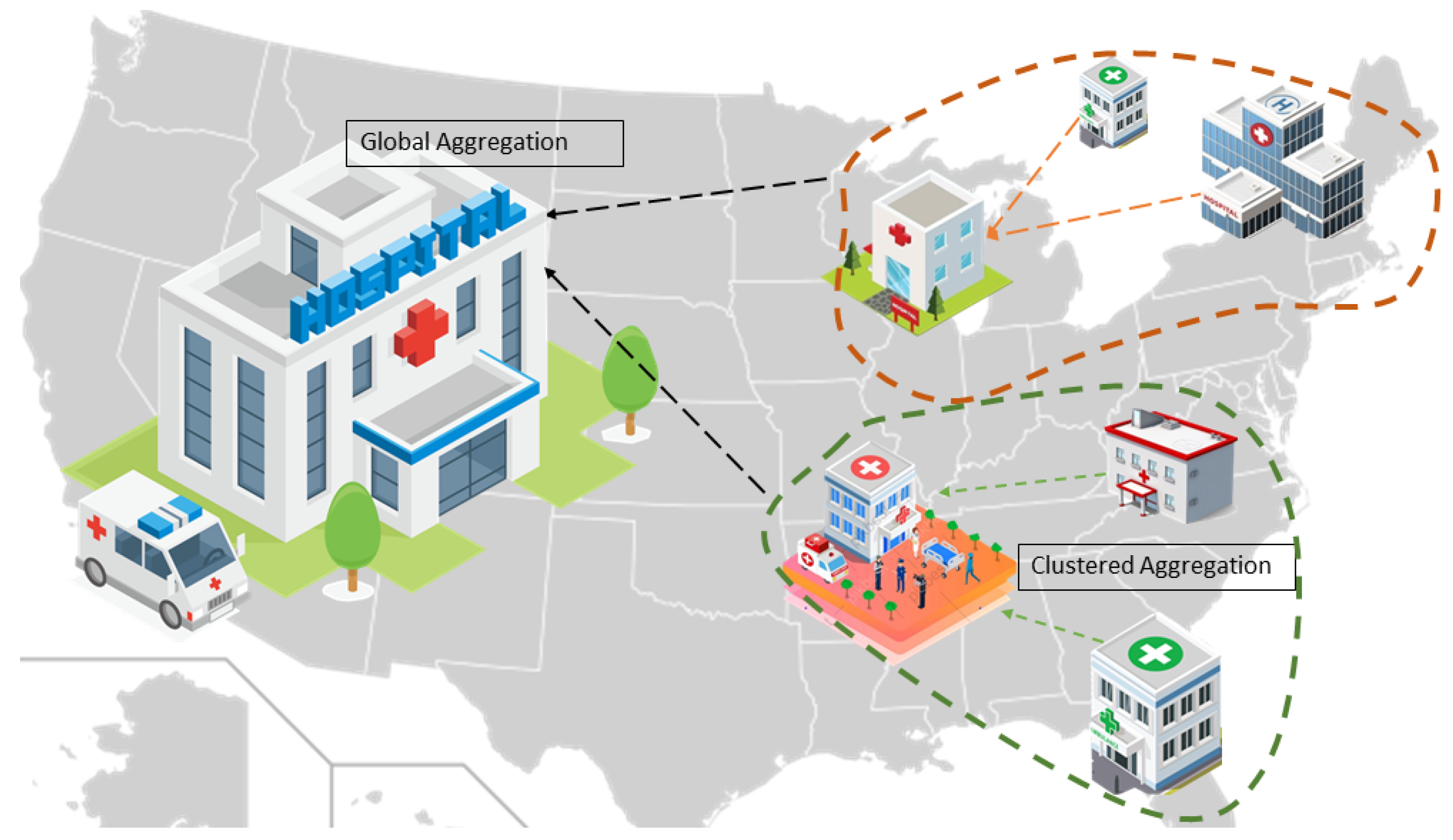
3. System Model
- : kth client’s dataset;
- : Total number of samples in patients’ data in the kth medical center;
- : Number of classes of labels in the kth medical center’s patients’ data;
- : Latent representation of the kth medical center’s patients’ data;
- : Global model at round t;
- : The kth medical center’s local model at round ;
- : Encoder network that maps input data to latent variable ;
- : The decoder network that maps latent variable ;
- : The divergence between latent spaces of client k and m;
- : Divergence threshold;
- : The mth group;
- : The kth client of the mth group;
- : Local model of the kth client of the mth group at round t;
- : Learning rate of the kth client of the mth group;
- : Gradients of the kth client of the mth group;
- : Clustered trained model of the mth group at round ;
- : Latent space of the mth group;
- : Latent space of the whole network;
- : Latent space of the kth client of the mth group;
- : The Kullback–Leibler divergence between learned latent distribution and prior distribution .
4. Proposed Model
4.1. Latent Representation Transformation
4.2. Group Formation
4.3. Client Update
| Algorithm 1 Proposed VAE based local model segmentation |
| 1: A. Initialization of Autoencoding: |
| 2: for do |
| 3: Latent representation using VAE of the kth client, |
| 4: |
| 5: end for |
| 6: for do |
| 7: for do |
| 8: Divergence evaluation using latent embedding of clients using Jensen–Shannon Divergence (JSD) |
| 9: |
| 10: with |
| 11: if then |
| 12: |
| 13: |
| 14: end if |
| 15: end for |
| 16: end for |
| 17: B. Initialization of Global Model Training: |
| Server execution: initialize |
| 18: for do |
| 19: for do |
| 20: for do |
| 21: |
| 22: end for |
| 23: |
| 24: |
| 25: end for |
| 26: |
| 27: |
| 28: end for |
4.4. Server Aggregation
5. Experiments
5.1. Data Set
5.2. Network Model
5.3. Results
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AP | Affinity propagation |
| CNN | Convolutional neural network |
| DNN | Deep neural network |
| FedAvg | Federated averaging |
| Non-IID | Non-independent and non-identically distributed |
| IoT | Internet of things |
| JSD | Jensen–Shanon divergence |
| MRI | Magnetic resonance imaging |
| VAE | Variational autoencoder |
References
- Mengistu, T.M.; Kim, T.; Lin, J.W. A Survey on Heterogeneity Taxonomy, Security and Privacy Preservation in the Integration of IoT, Wireless Sensor Networks and Federated Learning. Sensors 2024, 24, 968. [Google Scholar] [CrossRef] [PubMed]
- Hyysalo, J.; Dasanayake, S.; Hannu, J.; Schuss, C.; Rajanen, M.; Leppänen, T.; Doermann, D.; Sauvola, J. Smart mask—Wearable IoT solution for improved protection and personal health. IoT 2022, 18, 100511. [Google Scholar] [CrossRef] [PubMed]
- Avanzato, R.; Beritelli, F.; Lombardo, A.; Ricci, C. Lung-DT: An AI-Powered Digital Twin Framework for Thoracic Health Monitoring and Diagnosis. Sensors 2024, 24, 958. [Google Scholar] [CrossRef]
- Chaves, A.J.; Martín, C.; Díaz, M. Towards flexible data stream collaboration: Federated Learning in Kafka-ML. IoT 2024, 25, 101036. [Google Scholar] [CrossRef]
- Sun, P.; Shen, S.; Wan, Y.; Wu, Z.; Fang, Z.; Gao, X.-Z. A survey of iot privacy security: Architecture, technology, challenges, and trends. IEEE IoT J. 2024. [Google Scholar] [CrossRef]
- Bhatti, D.M.S.; Nam, H. FedCLS: Class-Aware Federated Learning in a Heterogeneous Environment. IEEE Trans. Netw. Serv. Manag. 2023, 20, 1517–1528. [Google Scholar] [CrossRef]
- Hossain, M.B.; Shinde, R.K.; Oh, S.; Kwon, K.C.; Kim, N. A Systematic Review and Identification of the Challenges of Deep Learning Techniques for Undersampled Magnetic Resonance Image Reconstruction. Sensors 2024, 24, 753. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.N.; Rizwan, A.; Ahmad, R.; Khan, Q.W.; Lim, S.; Kim, D.H. A precision-centric approach to overcoming data imbalance and non-IIDness in federated learning. IoT 2023, 23, 100890. [Google Scholar] [CrossRef]
- Bhatti, D.M.S.; Nam, H. A Performance Efficient Approach of Global Training in Federated Learning. In Proceedings of the 2023 International Conference on Artificial Intelligence in Information and Communication (ICAIIC), Bali, Indonesia, 20–23 February 2023; pp. 112–115. [Google Scholar] [CrossRef]
- Bhatti, D.M.S.; Saeed, N.; Nam, H. Fuzzy C-Means Clustering and Energy Efficient Cluster Head Selection for Cooperative Sensor Network. Sensors 2016, 16, 1459. [Google Scholar] [CrossRef]
- Sattler, F.; Müller, K.R.; Samek, W. Clustered Federated Learning: Model-Agnostic Distributed Multitask Optimization Under Privacy Constraints. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 3710–3722. [Google Scholar] [CrossRef]
- Ghosh, A.; Chung, J.; Yin, D.; Ramchandran, K. An Efficient Framework for Clustered Federated Learning. IEEE Trans. Inf. Theory 2022, 68, 8076–8091. [Google Scholar] [CrossRef]
- Briggs, C.; Fan, Z.; András, P. Federated learning with hierarchical clustering of local updates to improve training on non-IID data. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–9. [Google Scholar]
- Xie, M.K.; Long, G.; Shen, T.; Zhou, T.; Wang, X.; Jiang, J. Multi-Center Federated Learning. arXiv 2020, arXiv:2108.08647. [Google Scholar]
- Abad, M.S.H.; Ozfatura, E.; Gündüz, D.; Erçetin, Ö. Hierarchical Federated Learning ACROSS Heterogeneous Cellular Networks. In Proceedings of the ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 8866–8870. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Song, S.; Letaief, K.B. Client-Edge-Cloud Hierarchical Federated Learning. In Proceedings of the 2020 IEEE International Conference on Communications, Dublin, Ireland, 7–11 June 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Duan, M.; Liu, D.; Ji, X.; Liu, R.; Liang, L.; Chen, X.; Tan, Y. FedGroup: Ternary Cosine Similarity-based Clustered Federated Learning Framework toward High Accuracy in Heterogeneous Data. arXiv 2020, arXiv:2010.06870. [Google Scholar]
- Qiao, C.; Brown, K.N.; Zhang, F.; Tian, Z. Federated Adaptive Asynchronous Clustering Algorithm for Wireless Mesh Networks. IEEE Trans. Knowl. Data Eng. 2021, 35, 2610–2627. [Google Scholar] [CrossRef]
- Huang, Y.; Chu, L.; Zhou, Z.; Wang, L.; Liu, J.; Pei, J.; Zhang, Y. Personalized Federated Learning: An Attentive Collaboration Approach. arXiv 2020, arXiv:2007.03797. [Google Scholar]
- Qayyum, A.; Ahmad, K.; Ahsan, M.A.; Al-Fuqaha, A.I.; Qadir, J. Collaborative Federated Learning For Healthcare: Multi-Modal COVID-19 Diagnosis at the Edge. arXiv 2021, arXiv:2101.07511. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Song, S.; Letaief, K.B. Hierarchical Federated Learning with Quantization: Convergence Analysis and System Design. IEEE Trans. Wirel. Commun. 2022, 22, 2–18. [Google Scholar] [CrossRef]
- Luo, S.; Chen, X.; Wu, Q.; Zhou, Z.; Yu, S. HFEL: Joint Edge Association and Resource Allocation for Cost-Efficient Hierarchical Federated Edge Learning. IEEE Trans. Wirel. Commun. 2020, 19, 6535–6548. [Google Scholar] [CrossRef]
- Duan, M.; Liu, D.; Chen, X.; Tan, Y.; Ren, J.; Qiao, L.; Liang, L. Astraea: Self-Balancing Federated Learning for Improving Classification Accuracy of Mobile Deep Learning Applications. In Proceedings of the 2019 IEEE 37th International Conference on Computer Design (ICCD), Abu Dhabi, United Arab Emirates, 17–20 November 2019; pp. 246–254. [Google Scholar] [CrossRef]
- Bhatti, D.M.S.; Ahmed, S.; Chan, A.S.; Saleem, K. Clustering formation in cognitive radio networks using machine learning. AEU-Int. J. Electron. Commun. 2020, 114, 152994. [Google Scholar] [CrossRef]
- Dennis, D.K.; Li, T.; Smith, V. Heterogeneity for the Win: One-Shot Federated Clustering. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 2611–2620. [Google Scholar]
- Lloyd, S. Least squares quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Gholizadeh, N.; Musilek, P. Federated learning with hyperparameter-based clustering for electrical load forecasting. IoT 2022, 17, 100470. [Google Scholar] [CrossRef]
- de Moraes Sarmento, E.M.; Ribeiro, I.F.; Marciano, P.R.N.; Neris, Y.G.; de Oliveira Rocha, H.R.; Mota, V.F.S.; da Silva Villaça, R. Forecasting energy power consumption using federated learning in edge computing devices. IoT 2024, 25, 101050. [Google Scholar] [CrossRef]
- Cao, X.; Sun, G.; Yu, H.; Guizani, M. PerFED-GAN: Personalized Federated Learning via Generative Adversarial Networks. IEEE IoT J. 2022, 10, 3749–3762. [Google Scholar] [CrossRef]
- Yao, X.; Huang, T.; Zhang, R.; Li, R.; Sun, L. Federated Learning with Unbiased Gradient Aggregation and Controllable Meta Updating. arXiv 2019, arXiv:1910.08234. [Google Scholar]
- Zhang, L.; Shen, L.; Ding, L.; Tao, D.; Duan, L.Y. Fine-tuning Global Model via Data-Free Knowledge Distillation for Non-IID Federated Learning. arXiv 2022, arXiv:2203.09249. [Google Scholar] [CrossRef]
- Li, T.; Sahu, A.K.; Zaheer, M.; Sanjabi, M.; Talwalkar, A.; Smith, V. Federated Optimization in Heterogeneous Networks. arXiv 2018, arXiv:1812.06127. [Google Scholar] [CrossRef]
- Fallah, A.; Mokhtari, A.; Ozdaglar, A. Personalized Federated Learning: A Meta-Learning Approach. arXiv 2020, arXiv:2002.07948. [Google Scholar] [CrossRef]
- Bhatti, D.M.S.; Nam, H. A Robust Aggregation Approach for Heterogeneous Federated Learning. In Proceedings of the 2023 Fourteenth International Conference on Ubiquitous and Future Networks (ICUFN), Paris, France, 4–7 July 2023; pp. 300–304. [Google Scholar] [CrossRef]
- McMahan, H.B.; Moore, E.; Ramage, D.; Arcas, B.A.Y. Federated Learning of Deep Networks using Model Averaging. arXiv 2016, arXiv:1602.05629. [Google Scholar]
- Ahmed, T.; Longo, L. Examining the Size of the Latent Space of Convolutional Variational Autoencoders Trained With Spectral Topographic Maps of EEG Frequency Bands. IEEE Access 2022, 10, 107575–107586. [Google Scholar] [CrossRef]
- Sartaj, B.A.; Kadam, P.; Bhumkar, S.; Dedge, S.K. Brain Tumor Classification (MRI). 2020. Available online: https://www.kaggle.com/dsv/1183165 (accessed on 25 January 2024).
- Kermany, D.; Zhang, K.; Goldbaum, M. Labeled Optical Coherence Tomography and Chest X-ray Images for Classification. 2018. Available online: https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia/data (accessed on 28 February 2024).
- Xiao, H.; Rasul, K.; Vollgraf, R. Fashion-MNIST: A Novel Image Dataset for Benchmarking Machine Learning Algorithms. arXiv 2017, arXiv:1708.07747. [Google Scholar]
- Alex, K.; Vinod, N.; Geoffrey, H. The CIFAR-10 Dataset. 2009. Available online: http://www.cs.toronto.edu/~kriz/cifar.html (accessed on 25 November 2023).








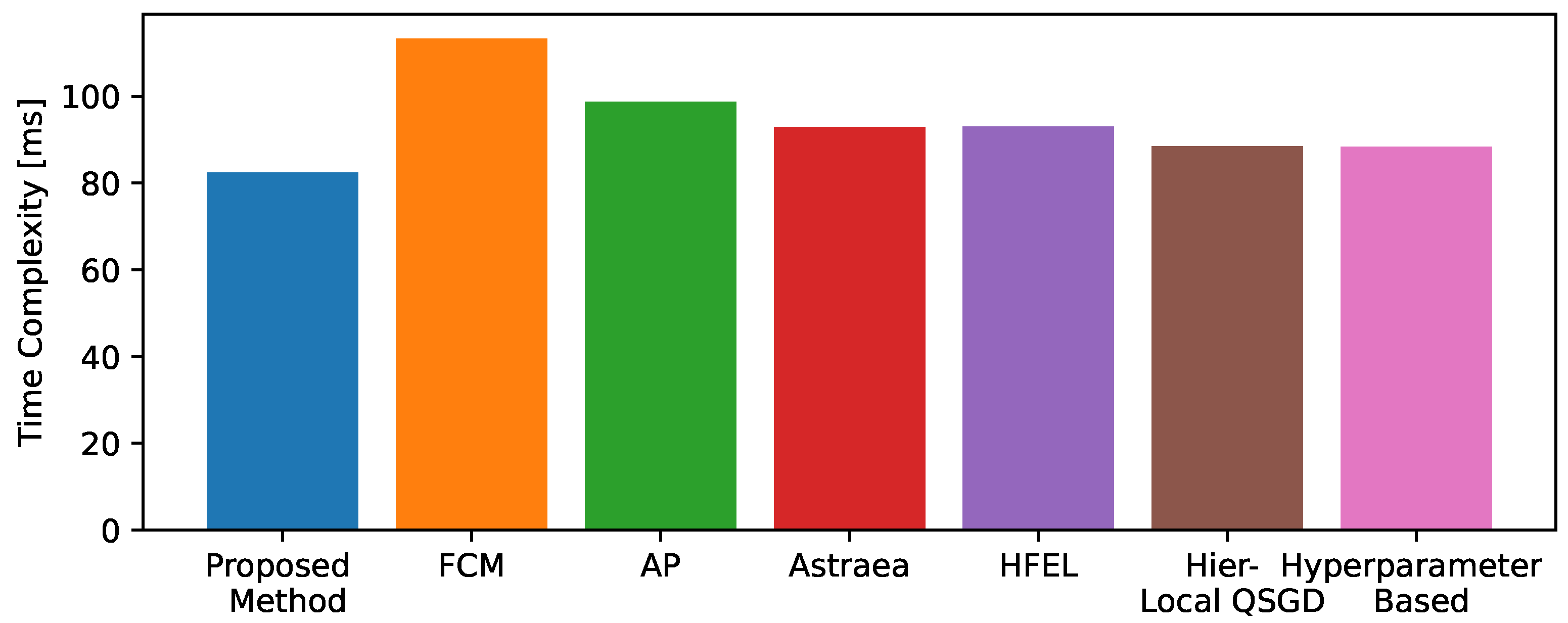
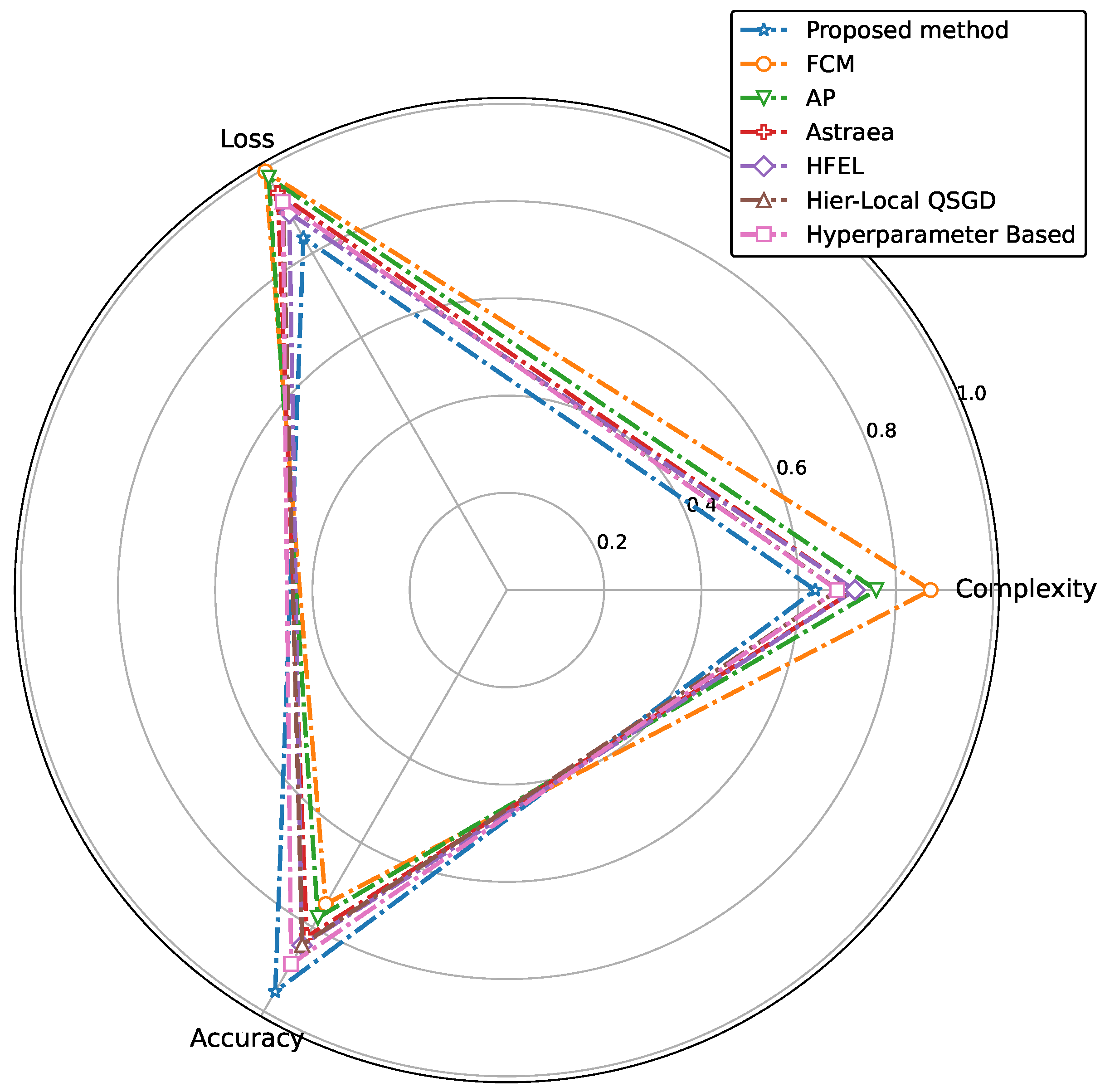
| Existing Methods | Framework Used | Clustering | Heterogeneity | Aggregation | Major Contributions |
|---|---|---|---|---|---|
| FCM [10] | Fuzzy c-means clustering | ✓ | × | × |
|
| Astraea [23] | Kullback–Leibler divergence (KLD) | × | ✓ | × |
|
| AP cluster formation [24] | Affinity propagation machine learning | ✓ | × | × |
|
| HFEL [22] | Hierarchical Federated Edge Learning | ✓ | × | ✓ |
|
| FL+ HC [13] | Distance-based clustering | ✓ | ✓ | × |
|
| k-FED [25] | Lloyd’s method for k-means clustering | ✓ | ✓ | × |
|
| Hier-Local QSGD [21] | Hierarchical Federated Learning with Quantization | ✓ | × | ✓ |
|
| Hyperparameter-Based [27] | Hyperparameter-based clustering | ✓ | × | × |
|
| Proposed method | Variational auto-encoder | ✓ | ✓ | ✓ |
|
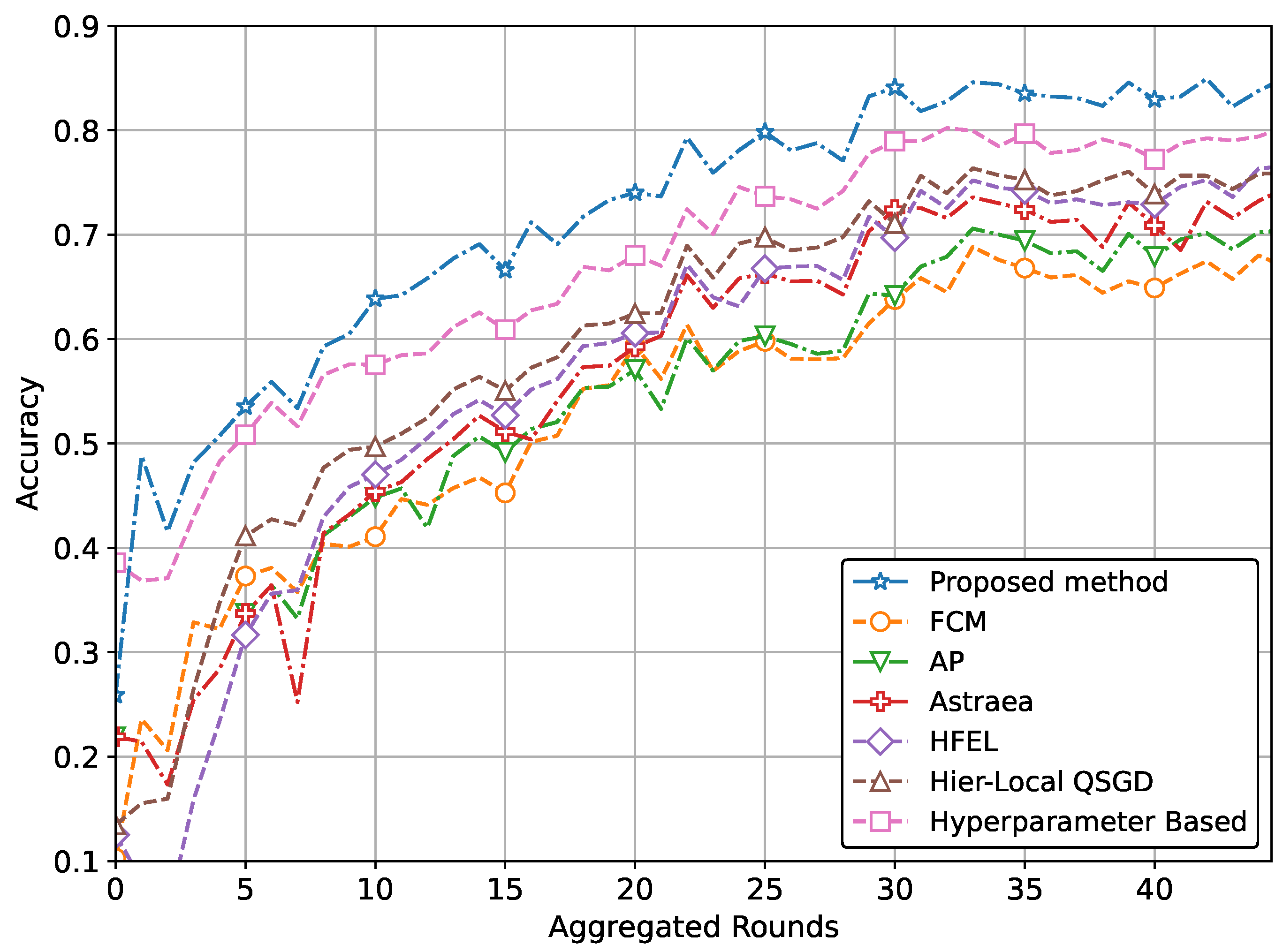
| (a) Performance with CNN network using MRI dataset | ||
| Comparing methods | Accuracy @MRI data | Loss @MRI data |
| FCM | 0.671 | 1.889 |
| AP | 0.700 | 1.862 |
| Astraea | 0.741 | 1.791 |
| HFEL | 0.759 | 1.701 |
| Hier-Local QSGD | 0.758 | 1.752 |
| Hyper-parameter Based | 0.799 | 1.752 |
| Proposed method | 0.848 | 1.675 |
| Performance improvement | 15% | 7% |
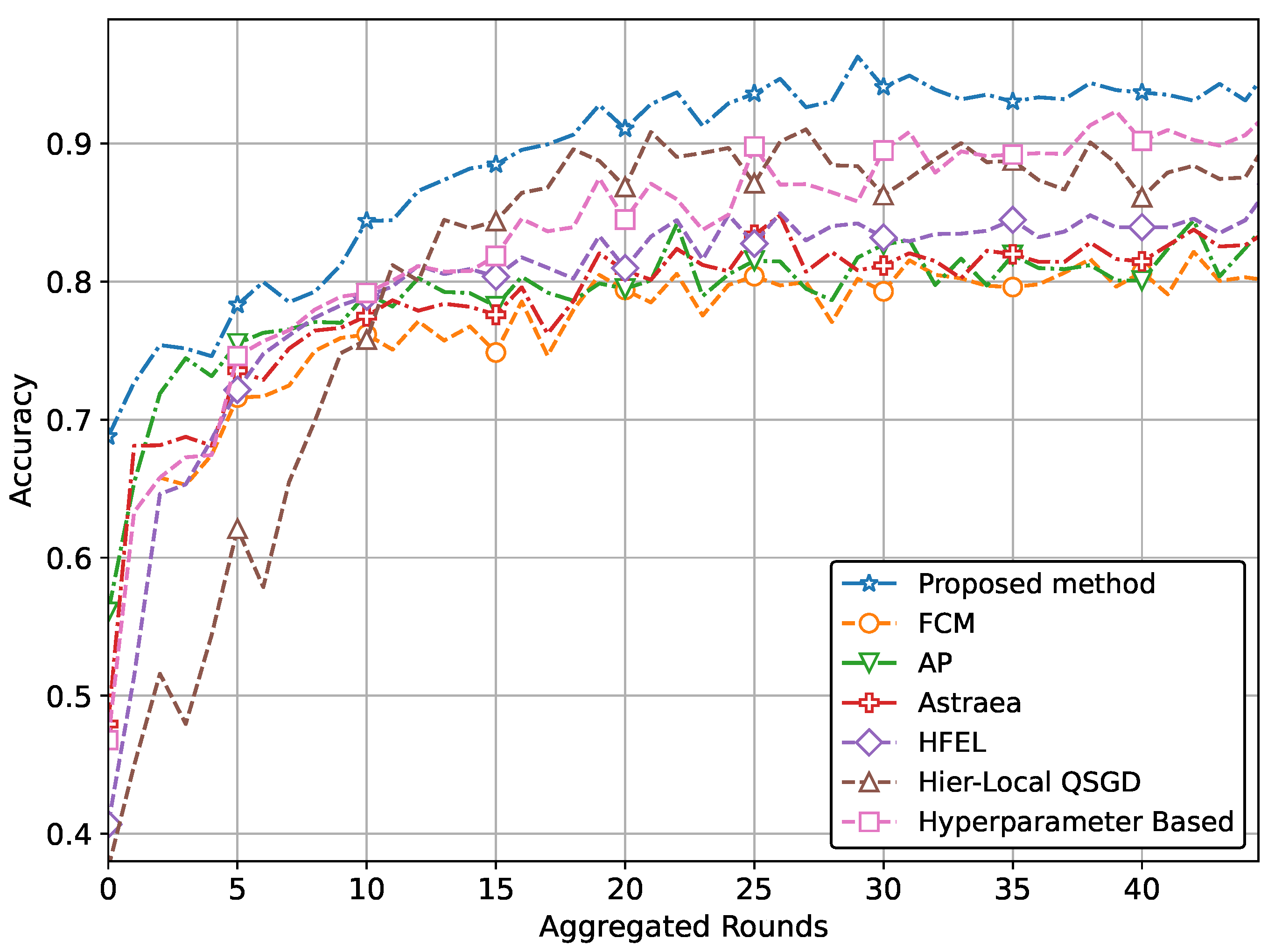
| (b) Performance with CNN network using pneumonia dataset | ||
| Comparing methods | Accuracy @pneumonia data | Loss @pneumonia data |
| FCM | 0.800 | 1.758 |
| AP | 0.830 | 1.711 |
| Astraea | 0.830 | 1.738 |
| HFEL | 0.849 | 1.701 |
| Hier-Local-QSGD | 0.891 | 1.668 |
| Hyper-parameter-based | 0.909 | 1.668 |
| Proposed method | 0.941 | 1.629 |
| Performance improvement | 10.5% | 4.8% |
| (c) Performance with DNN network using Fashion-MNIST | ||
| Comparing methods | Accuracy @Fashion MNIST | Loss @Fashion MNIST |
| FCM | 0.711 | 1.848 |
| AP | 0.716 | 1.819 |
| Astraea | 0.719 | 1.847 |
| HFEL | 0.741 | 1.801 |
| Hier-Local-QSGD | 0.771 | 1.779 |
| Hyper-parameter-based | 0.799 | 1.779 |
| Proposed method | 0.839 | 1.748 |
| Performance improvement | 13.1% | 3.7% |
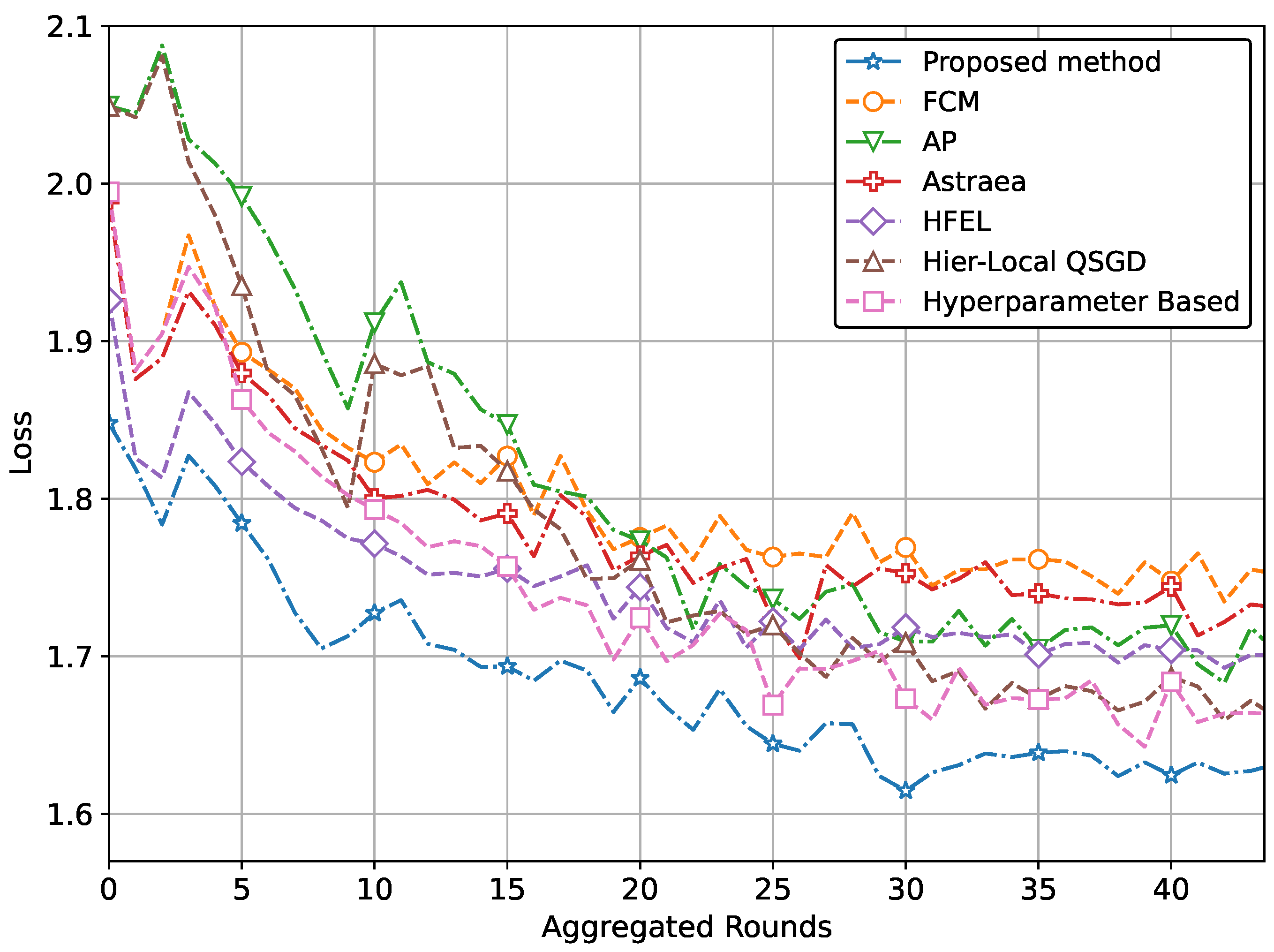
| (d) Performance with CNN network using CIFAR | ||
| Comparing methods | Accuracy @CIFAR | Loss @CIFAR |
| FCM | 0.448 | 2.079 |
| AP | 0.449 | 2.061 |
| Astraea | 0.450 | 2.066 |
| HFEL | 0.452 | 2.065 |
| Hier-Local-QSGD | 0.457 | 2.025 |
| Hyper-parameter-based | 0.485 | 2.026 |
| Proposed method | 0.551 | 1.960 |
| Performance improvement | 20.8% | 4.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhatti, D.M.S.; Choi, B.J. Enhancing IoT Healthcare with Federated Learning and Variational Autoencoder. Sensors 2024, 24, 3632. https://doi.org/10.3390/s24113632
Bhatti DMS, Choi BJ. Enhancing IoT Healthcare with Federated Learning and Variational Autoencoder. Sensors. 2024; 24(11):3632. https://doi.org/10.3390/s24113632
Chicago/Turabian StyleBhatti, Dost Muhammad Saqib, and Bong Jun Choi. 2024. "Enhancing IoT Healthcare with Federated Learning and Variational Autoencoder" Sensors 24, no. 11: 3632. https://doi.org/10.3390/s24113632
APA StyleBhatti, D. M. S., & Choi, B. J. (2024). Enhancing IoT Healthcare with Federated Learning and Variational Autoencoder. Sensors, 24(11), 3632. https://doi.org/10.3390/s24113632








