Calixarene-Based Supramolecular Sensor Array for Pesticide Discrimination
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Instruments
2.2. Competitive Fluorescence Titration
2.3. Discrimination of Different Kinds of Pesticides
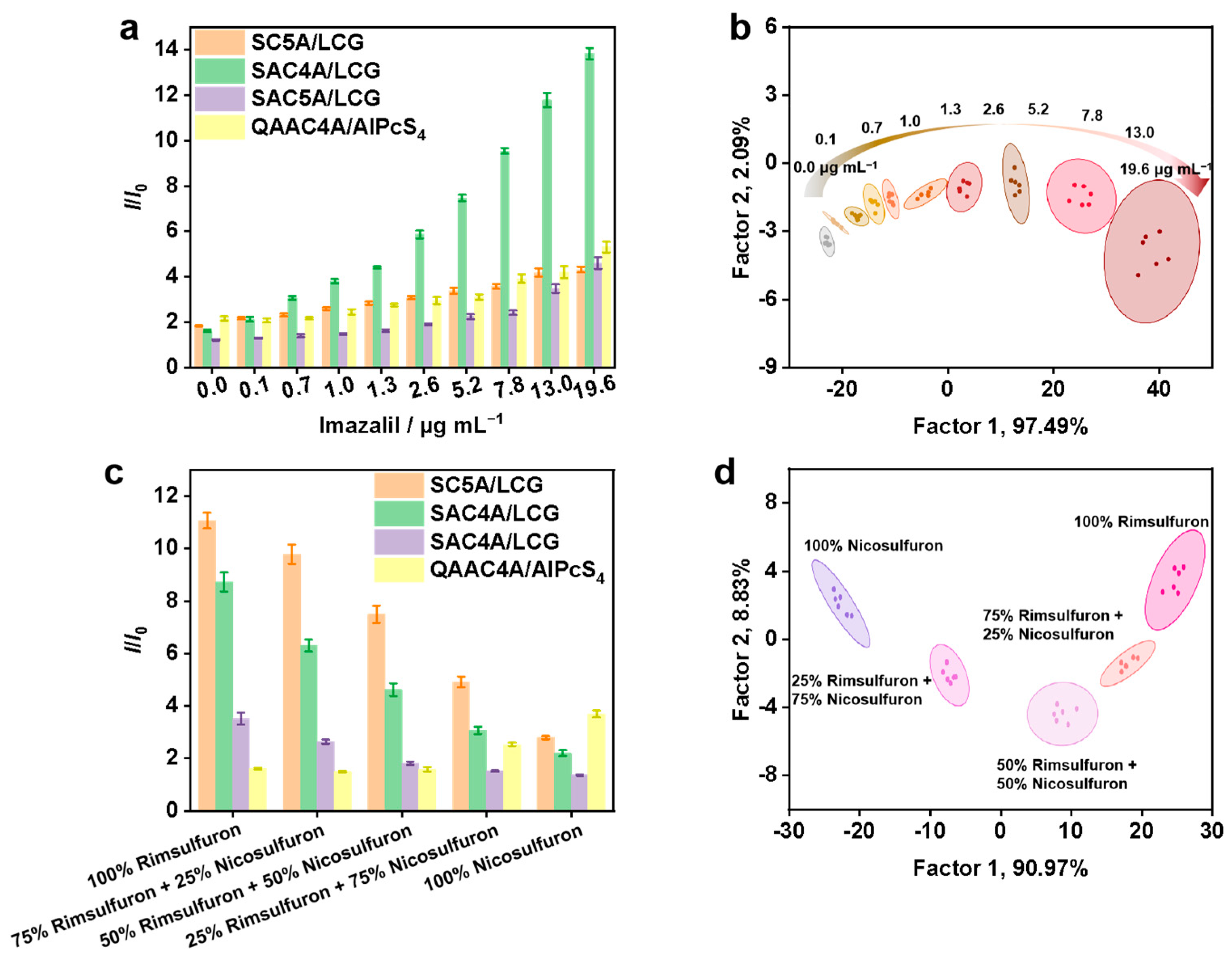
2.4. Discrimination of Different Concentrations of Imazalil and Bentazone
2.5. Discrimination of Mixed Pesticides
2.6. Visual Determination of Imazalil
3. Results and Discussion
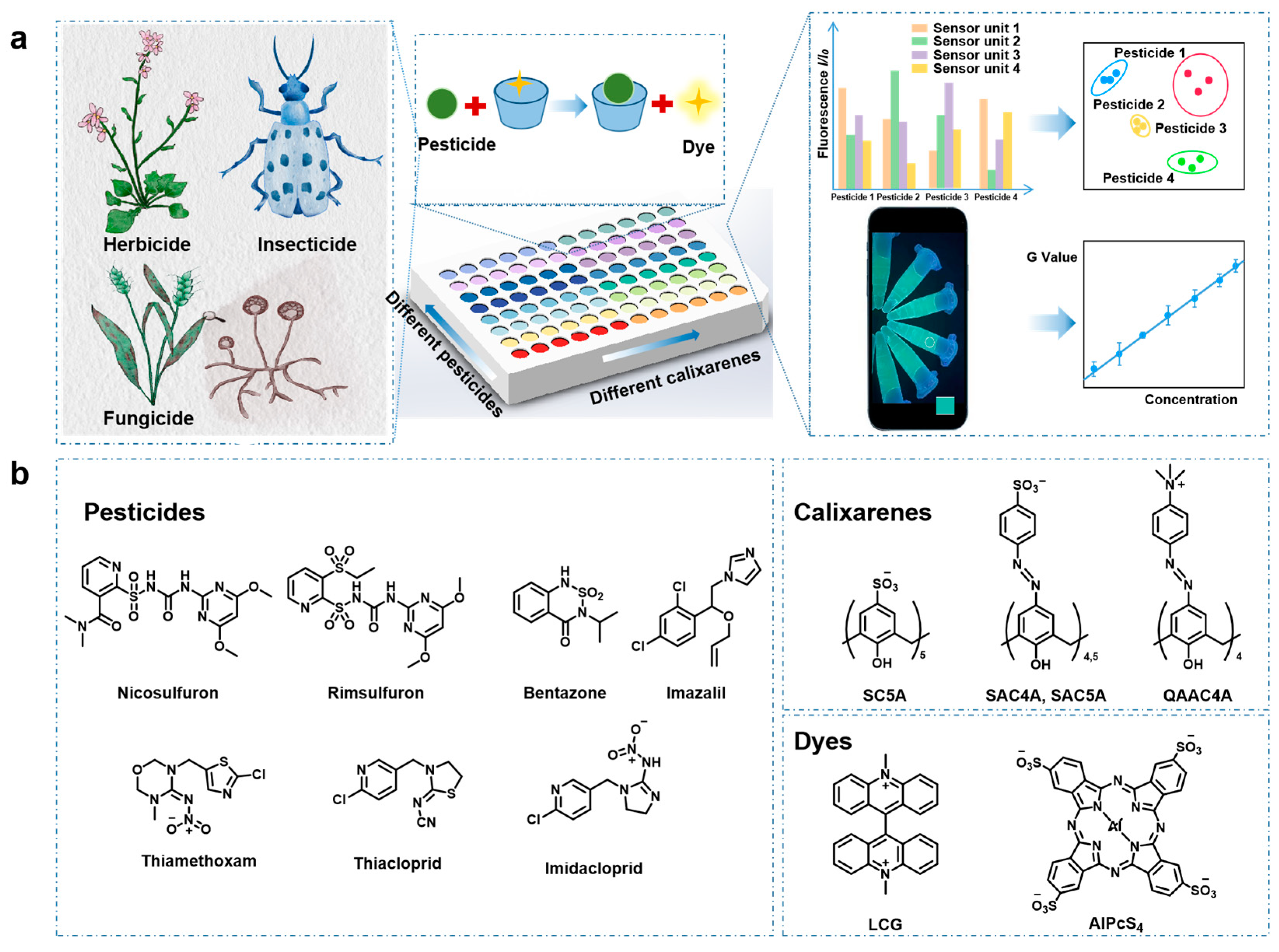
3.1. Principle of Supramolecular Sensor Array
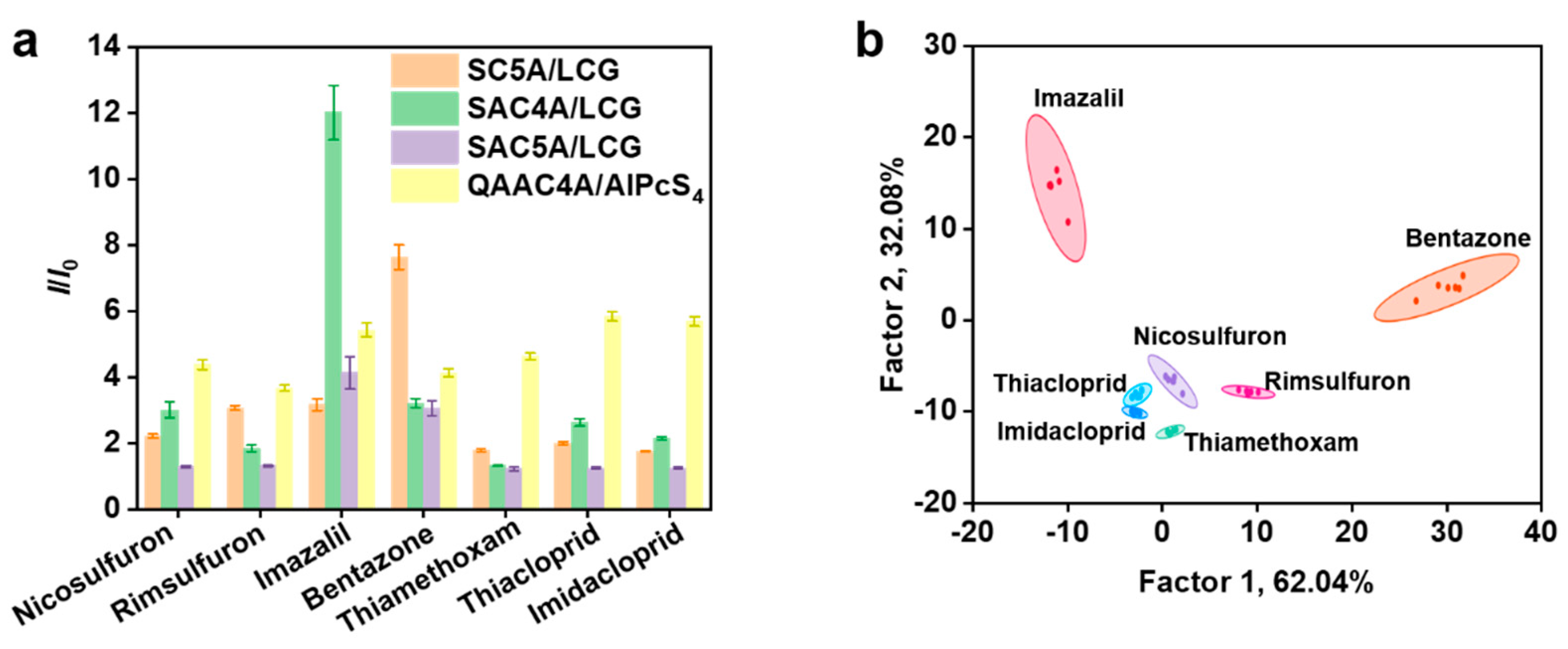
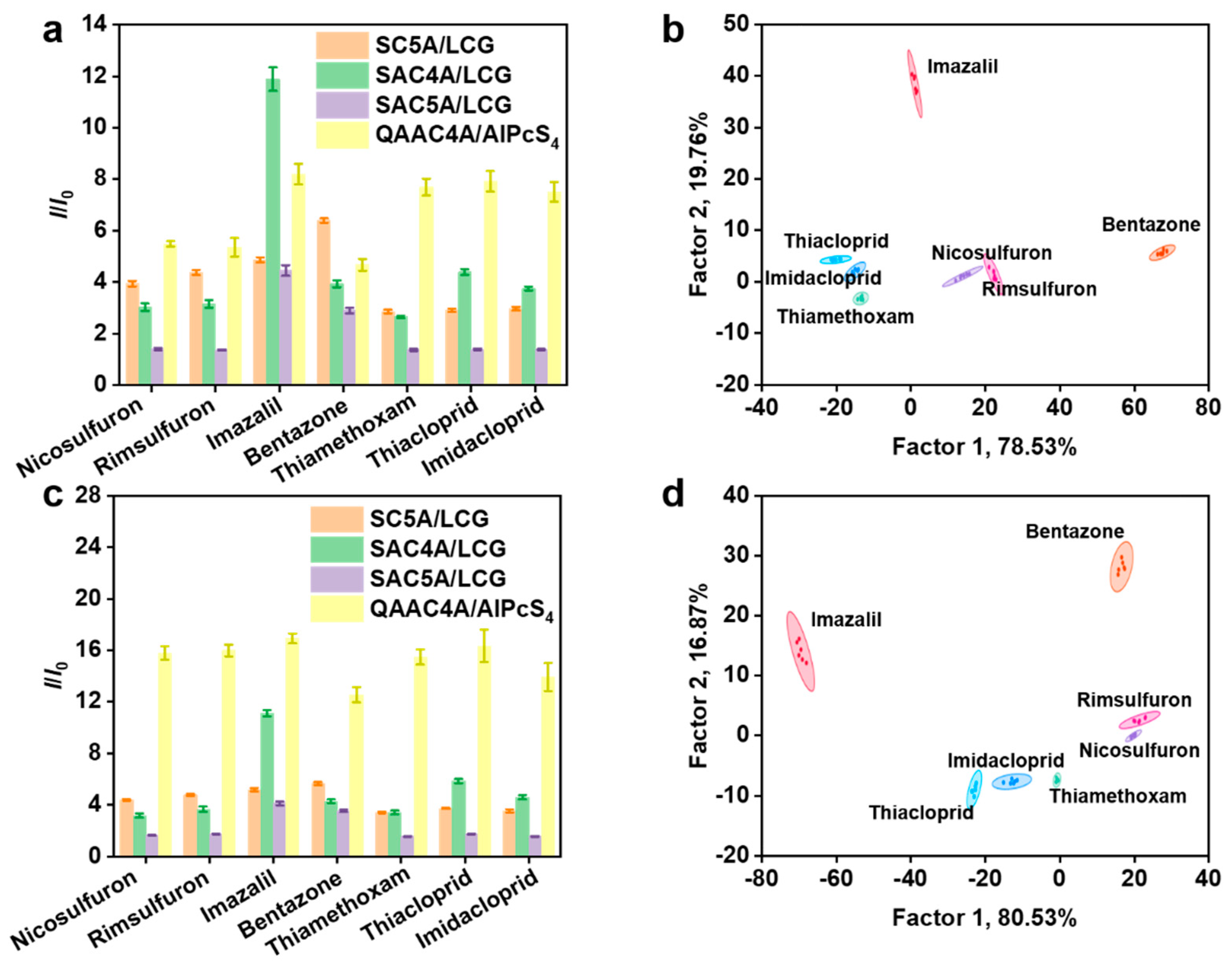
3.2. Discrimination of Pesticides by the Supramolecular Sensor Array
3.3. Real-Time Scanometric Monitoring of Imazalil
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, M.-L.; Gao, Y.; Han, X.X.; Zhao, B. Detection of pesticide residues in food using surface-enhanced Raman spectroscopy: A Review. J. Agric. Food Chem. 2017, 65, 6719–6726. [Google Scholar] [CrossRef] [PubMed]
- Golge, O.; Hepsag, F.; Kabak, B. Health risk assessment of selected pesticide residues in green pepper and cucumber. Food Chem. Toxicol. 2018, 121, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Choudhary, D.; Bali, S.; Bandral, S.S.; Singh, V.; Ahmad, M.A.; Rani, N.; Singh, T.G.; Chandrasekaran, B. Pesticides: An alarming detrimental to health and environment. Sci. Total Environ. 2024, 915, 170113. [Google Scholar] [CrossRef] [PubMed]
- Rani, L.; Thapa, K.; Kanojia, N.; Sharma, N.; Singh, S.; Grewal, A.S.; Srivastav, A.L.; Kaushal, J. An extensive review on the consequences of chemical pesticides on human health and environment. J. Clean. Prod. 2021, 283, 124657. [Google Scholar] [CrossRef]
- Aloizou, A.M.; Siokas, V.; Sapouni, E.M.; Sita, N.; Liampas, I.; Brotis, A.G.; Rakitskii, V.N.; Burykina, T.I.; Aschner, M.; Bogdanos, D.P.; et al. Parkinson’s disease and pesticides: Are microRNAs the missing link? Sci. Total Environ. 2020, 744, 140591. [Google Scholar] [CrossRef] [PubMed]
- Sabarwal, A.; Kumar, K.; Singh, R.P. Hazardous effects of chemical pesticides on human health-Cancer and other associated disorders. Environ. Toxicol. Pharmacol. 2018, 63, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiyama, T.; Katsuhara, M.; Nakajima, M. Compensation of matrix effects in gas chromatography-mass spectrometry analysis of pesticides using a combination of matrix matching and multiple isotopically labeled internal standards. J. Chromatogr. A 2017, 1524, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Maestroni, B.; Abu Alnaser, A.; Ghanem, I.; Islam, M.; Cesio, V.; Heinzen, H.; Kelly, S.; Cannavan, A. Validation of an analytical method for the determination of pesticide residues in vine leaves by GC-MS/MS. J. Agric. Food Chem. 2018, 66, 6421–6430. [Google Scholar] [CrossRef] [PubMed]
- García-Vara, M.; Postigo, C.; Palma, P.; Bleda, M.J.; Alda, M.L. QuEChERS-based analytical methods developed for LC-MS/MS multiresidue determination of pesticides in representative crop fatty matrices: Olives and sunflower seeds. Food Chem. 2022, 386, 132558. [Google Scholar] [CrossRef]
- Xiao, Z.; Yang, Y.; Li, Y.; Fan, X.; Ding, S. Determination of neonicotinoid insecticides residues in eels using subcritical water extraction and ultra-performance liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 2013, 777, 32–40. [Google Scholar] [CrossRef]
- Jiang, Y.; Sun, D.-W.; Pu, H.; Wei, Q. Surface enhanced Raman spectroscopy (SERS): A novel reliable technique for rapid detection of common harmful chemical residues. Trends Food Sci. Technol. 2018, 75, 10–22. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, L.; Zhang, Z.; Wu, J.; Zhang, L.; Jing, X.; Wang, X. Dispersive liquid–liquid microextraction combined with enzyme-linked immunosorbent assay for the analysis of chlorpyrifos in cereal samples. Talanta 2023, 265, 124802. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Fernández, B.; Mercader, J.V.; Checa-Orrego, B.I.; de la Escosura-Muñiz, A.; Costa-García, A. A monoclonal antibody-based immunosensor for the electrochemical detection of imidacloprid pesticide. Analyst 2019, 144, 2936–2941. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.-L.; Liu, Y.; Gu, H.-W.; Zhang, Q.; Zhang, Z.; Li, H. Multicolor enzyme-linked immunosorbent sensor for sensitive detection of organophosphorus pesticides based on TMB2+-mediated etching of gold nanorods. Microchem. J. 2021, 168, 106411. [Google Scholar] [CrossRef]
- Kadu, R.D.; Keri, R.S.; DH, N.; Budagumpi, S. State-of-the-art electrochemical sensors for quantitative detection of pesticides. Appl. Organomet. Chem. 2023, 37, e7097. [Google Scholar] [CrossRef]
- Guo, Z.; Liu, C.; Yang, R.; Dong, G.; Yang, Y.; Liu, H.; Wu, N. Detection of pesticide in water using two-dimensional fluorescence correlation spectroscopy and N-way partial least squares. Spectrochim. Acta Part A 2020, 229, 117981. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Tang, Y.; Liu, Y.; Tao, H.; Wu, Y. A novel colorimetric strategy for rapid detection of dimethoate residue in vegetables based on enhancing oxidase-mimicking catalytic activity of cube-shape Ag2O particles. Sens. Actuators B 2022, 361, 131720. [Google Scholar] [CrossRef]
- Shellaiah, M.; Sun, K.-W. Review on Anti-Aggregation-Enabled Colorimetric Sensing Applications of Gold and Silver Nanoparticles. Chemosensors 2022, 10, 536. [Google Scholar] [CrossRef]
- Leskovac, A.; Petrović, S. Pesticide Use and Degradation Strategies: Food Safety, Challenges and Perspectives. Foods 2023, 12, 2709. [Google Scholar] [CrossRef]
- Koushkestani, M.; Ghasemi, F.; Hormozi-Nezhad, M.R. Ratiometric Dual-Mode Optical Sensor Array for the Identification and Differentiation of Pesticides in Vegetables with Mixed Plasmonic and Fluorescent Nanostructures. ACS Appl. Nano Mater. 2024, 7, 2764–2774. [Google Scholar] [CrossRef]
- Song, D.; Tian, T.; Wang, L.; Zou, Y.; Zhao, L.; Xiao, J.; Huang, H.; Li, Y. Multi-signal sensor array based on a fluorescent nanozyme for broad-spectrum screening of pesticides. Chem. Eng. J. 2024, 482, 148784. [Google Scholar] [CrossRef]
- Tian, T.; Song, D.; Zhang, L.; Huang, H.; Li, Y. Facile and selective recognition of sulfonylurea pesticides based on the multienzyme-like activities enhancement of nanozymes combining sensor array. J. Hazard. Mater. 2024, 469, 133847. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Zha, D.; Anslyn, E.V. Recent advances in supramolecular analytical chemistry using optical sensing. Chem. Rev. 2015, 115, 7840–7892. [Google Scholar] [CrossRef] [PubMed]
- Albert, K.J.; Lewis, N.S.; Schauer, C.L.; Sotzing, G.A.; Stitzel, S.E.; Vaid, T.P.; Walt, D.R. Cross-reactive chemical sensor arrays. Chem. Rev. 2000, 100, 2595–2626. [Google Scholar] [CrossRef]
- Anslyn, E.V. Supramolecular analytical chemistry. J. Org. Chem. 2007, 72, 687–699. [Google Scholar] [CrossRef]
- Chen, F.-Y.; Li, C.-Z.; Han, H.; Geng, W.-C.; Zhang, S.-X.; Jiang, Z.-T.; Zhao, Q.-Y.; Cai, K.; Guo, D.-S. Expanding the hydrophobic cavity surface of azocalix[4]arene to enable biotin/avidin affinity with controlled release. Angew. Chem. Int. Ed. 2024, 136, e202402139. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.-X.; Lin, Y.-L.; Chen, M.-M.; Tian, H.-W.; Ma, R.; Wang, Z.-H.; Chen, F.-Y.; Pan, Y.-C.; Guo, D.-S. Azocalixarenes: A scaffold of universal excipients with high efficiency. Sci. China Chem. 2024, 67, 1697–1706. [Google Scholar] [CrossRef]
- Anzenbacher, P., Jr.; Lubal, P.; Buček, P.; Palacios, M.A.; Kozelkova, M.E. A practical approach to optical cross-reactive sensor arrays. Chem. Soc. Rev. 2010, 39, 3954–3979. [Google Scholar] [CrossRef]
- Zhang, C.; Yi, X.; Chen, C.; Tian, D.; Liu, H.; Xie, L.; Zhu, X.; Huang, M.; Ying, G.-G. Contamination of neonicotinoid insecticides in soil-water-sediment systems of the urban and rural areas in a rapidly developing region: Guangzhou, South China. Environ. Int. 2020, 139, 105719. [Google Scholar] [CrossRef]
- Jeschke, P.; Nauen, R.; Schindler, M.; Elbert, A. Overview of the status and global strategy for neonicotinoids. J. Agric. Food Chem. 2011, 59, 2897–2908. [Google Scholar] [CrossRef]
- Zhang, T.-X.; Zhang, Z.-Z.; Yue, Y.-X.; Hu, X.-Y.; Huang, F.; Shi, L.; Liu, Y.; Guo, D.-S. A general hypoxia-responsive molecular container for tumor-targeted therapy. Adv. Mater. 2020, 32, 1908435. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, M.; Shabbir, S.H.; Anslyn, E.V. Guidelines for pattern recognition using differential receptors and indicator displacement assays. J. Org. Chem. 2009, 74, 4479–4489. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.-H.; Hu, X.-Y.; Hu, Z.-Y.; Tian, H.-W.; Li, J.-J.; Pan, Y.-C.; Li, H.-B.; Guo, D.-S. A facile way to construct sensor array library via supramolecular chemistry for discriminating complex systems. Nat. Commun. 2022, 13, 4293. [Google Scholar] [CrossRef] [PubMed]
- Fargher, H.A.; d’Oelsnitz, S.; Diaz, D.J.; Anslyn, E.V. Pushing differential sensing further: The next steps in design and analysis of bio-inspired cross-reactive arrays. Anal. Sens. 2023, 3, e202200095. [Google Scholar] [CrossRef]
- Askim, J.R.; Mahmoudi, M.; Suslick, K.S. Optical sensor arrays for chemical sensing: The optoelectronic nose. Chem. Soc. Rev. 2013, 42, 8649–8682. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-H.; Kabir, E.; Jahan, S.A. Exposure to pesticides and the associated human health effects. Sci. Total Environ. 2017, 575, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Uclés, A.; García, A.V.; Gil García, M.D.; Aguilera del Real, A.M.; Fernández-Alba, A.R. Benzimidazole and imidazole fungicide analysis in grape and wine samples using a competitive enzyme-linked immunosorbent assay. Anal. Methods 2015, 7, 9158–9165. [Google Scholar] [CrossRef]
- Gutsche, C.D.; Bauer, L.J. Calixarenes. 13. The conformational properties of calix[4]arenes, calix[6]arenes, calix[8]arenes, and oxacalixarenes. J. Am. Chem. Soc. 1985, 107, 6052–6059. [Google Scholar] [CrossRef]
- Lu, L.; Zhu, S.; Liu, X.; Xie, Z.; Yan, X. Highly selective chromogenic ionophores for the recognition of chromium(III) based on a water-soluble azocalixarene derivative. Anal. Chim. Acta 2005, 535, 183–187. [Google Scholar] [CrossRef]
- Yue, Y.-X.; Zhang, Z.; Wang, Z.-H.; Ma, R.; Chen, M.-M.; Ding, F.; Li, H.-B.; Li, J.-J.; Shi, L.; Liu, Y.; et al. Promoting tumor accumulation of anticancer drugs by hierarchical carrying of exogenous and endogenous vehicles. Small Struct. 2022, 3, 2200067. [Google Scholar] [CrossRef]
- Shinkai, S.; Araki, K.; Shibata, J.; Tsugawa, D.; Manabe, O. Diazo-coupling reactions with calix[4]arene. pKa determination with chromophoric azocalix[4]arenes. Chem. Lett. 1989, 18, 931–934. [Google Scholar] [CrossRef]
- He, Y.; Xu, B.; Li, W.; Yu, H. Silver nanoparticle-based chemiluminescent sensor array for pesticide discrimination. J. Agric. Food Chem. 2015, 63, 2930–2934. [Google Scholar] [CrossRef] [PubMed]
- Fahimi-Kashani, N.; Hormozi-Nezhad, M.R. Gold-nanoparticle-based colorimetric sensor array for discrimination of organophosphate pesticides. Anal. Chem. 2016, 88, 8099–8106. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.-N.; Yang, R.-P.; Huang, S.-Z.; Tao, Z.; Tang, Q.; Huang, Y. Supramolecular fluorescence sensor array based on cucurbit[8]uril complexes used for the detection of multiplex quaternary ammonium pesticides. J. Agric. Food Chem. 2023, 71, 9549–9557. [Google Scholar] [CrossRef]
- Kumar, M.; Kaur, N.; Singh, N. Colorimetric nanozyme sensor array based on metal nanoparticle-decorated CNTs for quantification of pesticides in real water and soil samples. ACS Sustainable Chem. Eng. 2024, 12, 728–736. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Tian, J.-H.; Tian, H.-W.; Ma, R.; Wang, Z.-H.; Pan, Y.-C.; Hu, X.-Y.; Guo, D.-S. Calixarene-Based Supramolecular Sensor Array for Pesticide Discrimination. Sensors 2024, 24, 3743. https://doi.org/10.3390/s24123743
Chen Y, Tian J-H, Tian H-W, Ma R, Wang Z-H, Pan Y-C, Hu X-Y, Guo D-S. Calixarene-Based Supramolecular Sensor Array for Pesticide Discrimination. Sensors. 2024; 24(12):3743. https://doi.org/10.3390/s24123743
Chicago/Turabian StyleChen, Yeye, Jia-Hong Tian, Han-Wen Tian, Rong Ma, Ze-Han Wang, Yu-Chen Pan, Xin-Yue Hu, and Dong-Sheng Guo. 2024. "Calixarene-Based Supramolecular Sensor Array for Pesticide Discrimination" Sensors 24, no. 12: 3743. https://doi.org/10.3390/s24123743



