Next-Gen Medical Imaging: U-Net Evolution and the Rise of Transformers
Abstract
1. Introduction
2. Medical Imaging Segmentation
2.1. Difficulty from Imaging Sensor
- High resolution: the number of pixels of the sensor is crucial, as a higher resolution allows for more detailed images, which is essential for accurate diagnosis;
- High sensitivity: the sensor’s performance in low-light conditions ensures that high sensitivity provides clear images even with low radiation doses, enhancing patient safety;
- High noise level: the random electrical signals generated during image capture need to be minimized since lower noise levels lead to clearer and more accurate images, reducing the likelihood of misdiagnosis.
2.2. U-Net and Its Variants’ Structures
2.2.1. Residual Module
2.2.2. Intention Module
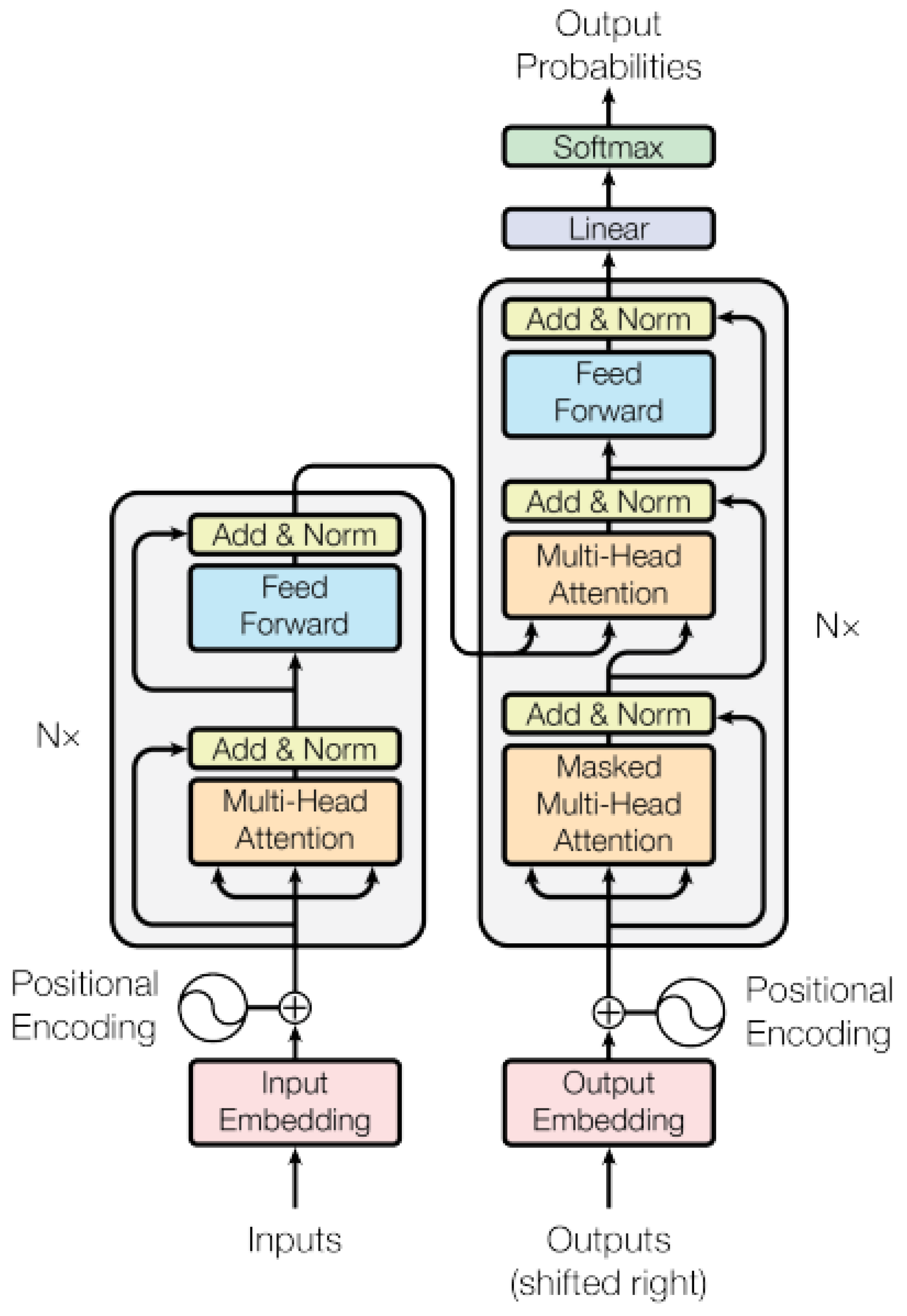
3. Transformer in Medical Imaging
- The algorithm will initialize the matrix for K, Q, and V.
- The relationship between Queries and Keys: Each Query must first be connected to every Key. First, to facilitate the dot product operation, the algorithm transposes the K matrix and then multiplies the transposed matrix by the Q matrix. This indicates that each Query computes its correlation with each Key to determine which Keys are more pertinent to a given Query.
- Scaling: The results of the correlation calculations are ordinarily divided by the square root of dk () to ensure consistent computations. This step helps to control the range of values to ensure the stability of the calculation.
- Softmax: The Softmax function is applied to convert the correlation distribution of Query to Keys into a weight distribution. The Softmax function ensures that the sum of these weights is equal to 1, and the appropriate weights are assigned according to the strength of the correlation.
- Multiplication of Weights with Values: These weights are multiplied by the corresponding values. This step weights the Query information with the related Key details to produce the final output.
3.1. Transformers in Computer Vision
Self-Attention and Convolutional Operation
3.2. Feature Extraction
3.2.1. Hybrid Structure
3.2.2. Self-Attention Block
- W-MSA: In W-MSA, input data are divided into windows, each containing multiple adjacent blocks of data. Self-attention operations are performed in each window, allowing each block of data to perform self-attention calculations with other blocks in the same window. This helps capture local features.
- SW-MSA: SW-MSA is an improved multi-head self-attention mechanism that introduces the offset of the window. This means that when calculating self-attention, it is no longer limited to the data blocks within the window but takes into account the relationships between the windows. This helps capture a wider range of contextual information.

- Transformer position selection, which affects model performance: Choosing a segmentation model that places the Transformer in the encoder is more common than a segmentation model that places it in the decoder. This is because encoders are mainly used to extract features, while decoders are used primarily to fuse features extracted by encoders.
- Feature expression ability improvement: In order to better fuse global and local information, it is common to use a Transformer in the encoder to extract information and then use a Transformer in the decoder to fuse the information and combine the convolutional network to obtain detailed features as an advantage, so as to enhance the model’s ability to express features.
- Complexity and efficiency trade-offs: Inserting Transformer modules into both the encoder and decoder increases the computational complexity of the attention mechanism, resulting in a decrease in model efficiency. Therefore, efficient attention modules need to be explored to improve the efficiency of such models.
- Balance at transition junctions: Placing the Transformer at the transition junction is a trade-off option to draw connections from features with low expressiveness while relying on global features to guide subsequent fusions. This is because the feature map at the transition junction has the lowest resolution, and even if you use a multi-layer superimposed Transformer module, it will not put a large load on the model. However, this approach has limited capabilities in feature extraction and fusion, and there is a trade-off between its use and its costs.
3.2.3. Others
3.2.4. Summary
3.3. Learning Strategy
3.3.1. Semi-Supervision
3.3.2. Class Awareness Enhancement
3.3.3. Uncertainty Awareness
4. Discussion and Limitation
- Data limitations: Medical image datasets are more difficult to obtain than ordinary computer vision datasets [4]. The issues involved are more complex, including privacy concerns, data scarcity, diversity (such as X-rays, MRIs, CTs, ultrasounds, etc.), and the specialization of the medical field (which usually requires annotation by professional doctors). This data limitation poses a significant challenge for Transformer-based models, as they heavily rely on the self-attention mechanism to capture long-range dependencies and global context information [114]. The self-attention algorithm’s complexity scales quadratically with the input sequence length, making it computationally expensive, especially for high-resolution medical images. Consequently, Transformer-based models require larger datasets to learn the intricate patterns and relationships within medical images [116]. However, the scarcity of annotated medical data can hinder the model’s ability to leverage the self-attention mechanism fully, potentially limiting its performance compared to that of CNN, which is more parameter-efficient and can better generalize from smaller datasets.
- Generalization: Generalization is a prevalent concept in developing deep learning, particularly within computer vision, where large pre-trained models are commonplace [117]. These models are characterized by their extensive parameter count and intricate architecture. Among these, Transformer-based large models stand out as a prime example. They can adapt to various datasets within their respective domains with minimal effort, necessitating only fine-tuning for different applications [118,119]. This flexibility enables seamless migration from one task to another, eliminating the need for excessive additional training. However, the medical field faces a unique challenge in adopting pre-trained large models. This is primarily attributed to the intricacies and lack of medical data, making developing such models a formidable endeavor.
5. Conclusions and Research Direction
- Multi-scale feature extraction: In deep U-shaped networks, the upper model first learns broader features such as edges and textures, and as the network increases, the underlying structure extracts higher-level features. Then, at this time, the data are transferred between different levels, and data loss will inevitably occur [120]. This is also the reason why the model is less effective for segmentation. However, the previous model also has techniques such as regional feature enhancement or hierarchical feature jumping [72,80,83], although further research must emphasize enhancing edge detection and noise cancellation. One of the position directions could be federal learning, combining different reception fields to generate a more comprehensive result [121].
- Further local–global context extraction: To further enhance local and global information extraction, integrating advanced methods such as hybrid models that combine CNNs with Transformers can be promising. These models can leverage the strengths of CNNs in capturing fine-grained local features and the capability of Transformers in modeling long-range dependencies [122]. Additionally, incorporating multi-head self-attention mechanisms and hierarchical attention structures can improve the model’s ability to capture nuanced details and broader contextual information simultaneously [41]. Techniques such as attention gating can also selectively focus on relevant parts of the image, enhancing the overall segmentation accuracy [29]. Moreover, combining these methods with advanced data augmentation techniques and synthetic data generation can address the data scarcity issue and further improve the robustness and generalization of the models in medical image segmentation.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cheung, C.W.J.; Zhou, G.Q.; Law, S.Y.; Mak, T.M.; Lai, K.L.; Zheng, Y.P. Ultrasound volume projection imaging for assessment of scoliosis. IEEE Trans. Med. Imaging 2015, 34, 1760–1768. [Google Scholar] [CrossRef] [PubMed]
- Khademi, Z.; Ebrahimi, F.; Kordy, H.M. A review of critical challenges in MI-BCI: From conventional to deep learning methods. J. Neurosci. Methods 2023, 383, 109736. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Lyu, J.; Huang, Z.; Leung, F.H.; Lee, T.; Yang, D.; Su, S.; Zheng, Y.; Ling, S.H. Ultrasound spine image segmentation using multi-scale feature fusion Skip-Inception U-Net (SIU-Net). Biocybern. Biomed. Eng. 2022, 42, 341–361. [Google Scholar] [CrossRef]
- Willemink, M.J.; Koszek, W.A.; Hardell, C.; Wu, J.; Fleischmann, D.; Harvey, H.; Folio, L.R.; Summers, R.M.; Rubin, D.L.; Lungren, M.P. Preparing medical imaging data for machine learning. Radiology 2020, 295, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhang, J.; Xia, Y.; Wu, Q. Unified 2d and 3d pre-training for medical image classification and segmentation. arXiv 2021, arXiv:2112.09356. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015, Proceedings, Part III 18; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Aung, K.P.P.; Nwe, K.H. Regions of Interest (ROI) Analysis for Upper Limbs EEG Neuroimaging Schemes. In Proceedings of the 2020 International Conference on Advanced Information Technologies (ICAIT), Yangon, Myanmar, 4–5 November 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 53–58. [Google Scholar]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-net and its variants for medical image segmentation: A review of theory and applications. IEEE Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Steinegger, A.; Wolfbeis, O.S.; Borisov, S.M. Optical sensing and imaging of pH values: Spectroscopies, materials, and applications. Chem. Rev. 2020, 120, 12357–12489. [Google Scholar] [CrossRef] [PubMed]
- Westerveld, W.J.; Mahmud-Ul-Hasan, M.; Shnaiderman, R.; Ntziachristos, V.; Rottenberg, X.; Severi, S.; Rochus, V. Sensitive, small, broadband and scalable optomechanical ultrasound sensor in silicon photonics. Nat. Photonics 2021, 15, 341–345. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, N.; Yang, H.; Sun, J.; Xu, Z. Model-driven deep attention network for ultra-fast compressive sensing MRI guided by cross-contrast MR image. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020, Proceedings, Part II 23; Springer: Berlin/Heidelberg, Germany, 2020; pp. 188–198. [Google Scholar]
- Danielsson, M.; Persson, M.; Sjölin, M. Photon-counting x-ray detectors for CT. Phys. Med. Biol. 2021, 66, 03TR01. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, X.; Tian, N.; Liu, M.; Cai, Z.; Feng, P.; Dou, R.; Yu, S.; Wu, N.; Liu, J.; et al. A 64 × 128 3D-Stacked SPAD Image Sensor for Low-Light Imaging. Sensors 2024, 24, 4358. [Google Scholar] [CrossRef] [PubMed]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 1–74. [Google Scholar]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical image analysis using convolutional neural networks: A review. J. Med. Syst. 2018, 42, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, M.A.; Ling, S.H. Evolving optimised convolutional neural networks for lung cancer classification. Signals 2022, 3, 284–295. [Google Scholar] [CrossRef]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Moutik, O.; Sekkat, H.; Tigani, S.; Chehri, A.; Saadane, R.; Tchakoucht, T.A.; Paul, A. Convolutional neural networks or vision transformers: Who will win the race for action recognitions in visual data? Sensors 2023, 23, 734. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, M.A.; Ling, S.S.H.; Wong, J.K.W. Exploring the Frontier: Transformer-Based Models in EEG Signal Analysis for Brain-Computer Interfaces. Comput. Biol. Med. 2024, 178, 108705. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Zhang, D.; Liu, J.; Cheng, C. MSU-Net: Multi-scale U-Net for 2D medical image segmentation. Front. Genet. 2021, 12, 639930. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Z.; Wang, Y.; Huang, Y.H.; Xiang, P.; Liu, W.X.; Lai, Q.Q.; Gao, Y.Y.; Xu, M.S.; Guo, Y.F. RSU-Net: U-net based on residual and self-attention mechanism in the segmentation of cardiac magnetic resonance images. Comput. Methods Programs Biomed. 2023, 231, 107437. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, Q.; Wang, Y. Road extraction by deep residual u-net. IEEE Geosci. Remote. Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Johansen, D.; De Lange, T.; Halvorsen, P.; Johansen, H.D. Resunet++: An advanced architecture for medical image segmentation. In Proceedings of the 2019 IEEE International Symposium on Multimedia (ISM), San Diego, CA, USA, 9–11 December 2019; pp. 225–2255. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Wei, J.; Sun, B.; Su, S.; Zuo, Z.; Wu, P. ASCU-Net: Attention gate, spatial and channel attention u-net for skin lesion segmentation. Diagnostics 2021, 11, 501. [Google Scholar] [CrossRef]
- Khanh, T.L.B.; Dao, D.P.; Ho, N.H.; Yang, H.J.; Baek, E.T.; Lee, G.; Kim, S.H.; Yoo, S.B. Enhancing U-Net with spatial-channel attention gate for abnormal tissue segmentation in medical imaging. Appl. Sci. 2020, 10, 5729. [Google Scholar] [CrossRef]
- Li, C.; Tan, Y.; Chen, W.; Luo, X.; Gao, Y.; Jia, X.; Wang, Z. Attention unet++: A nested attention-aware u-net for liver ct image segmentation. In Proceedings of the 2020 IEEE International Conference on Image Processing (ICIP), Abu Dhabi, United Arab Emirates, 25–28 October 2020; pp. 345–349. [Google Scholar]
- Qiao, Z.; Du, C. Rad-unet: A residual, attention-based, dense unet for CT sparse reconstruction. J. Digit. Imaging 2022, 35, 1748–1758. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Lyu, J.; Huang, Z.; Leung, H.F.F.; Lee, T.T.Y.; Yang, D.; Su, S.; Zheng, Y.; Ling, S.H. Light-convolution Dense selection U-net (LDS U-net) for ultrasound lateral bony feature segmentation. Appl. Sci. 2021, 11, 10180. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, C.; Zhou, T.; Feng, L.; Liu, L.; Zeng, Q.; Wang, G. A deep residual attention-based U-Net with a biplane joint method for liver segmentation from CT scans. Comput. Biol. Med. 2023, 152, 106421. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, C.; Coleman, S.; Kerr, D. DENSE-INception U-net for medical image segmentation. Comput. Methods Programs Biomed. 2020, 192, 105395. [Google Scholar] [CrossRef]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef]
- McHugh, H.; Talou, G.M.; Wang, A. 2d Dense-UNet: A clinically valid approach to automated glioma segmentation. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 4 October 2020, Revised Selected Papers, Part II 6; Springer: Berlin/Heidelberg, Germany, 2021; pp. 69–80. [Google Scholar]
- Zhao, H.; Jia, J.; Koltun, V. Exploring self-attention for image recognition. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10076–10085. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. Transformers: State-of-the-art natural language processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, Online, 16–20 November 2020; pp. 38–45. [Google Scholar]
- Grigsby, J.; Wang, Z.; Nguyen, N.; Qi, Y. Long-range transformers for dynamic spatiotemporal forecasting. arXiv 2021, arXiv:2109.12218. [Google Scholar]
- Lund, B.D.; Wang, T. Chatting about ChatGPT: How may AI and GPT impact academia and libraries? Libr. Hi Tech News 2023, 40, 26–29. [Google Scholar] [CrossRef]
- Nadkarni, P.M.; Ohno-Machado, L.; Chapman, W.W. Natural language processing: An introduction. J. Am. Med. Inform. Assoc. 2011, 18, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.H.; Tiels, K.; Aguirre, L.A.; Schön, T. Beyond exploding and vanishing gradients: Analysing RNN training using attractors and smoothness. PMLR 2020, 108, 2370–2380. [Google Scholar]
- Fernández, S.; Graves, A.; Schmidhuber, J. Sequence labelling in structured domains with hierarchical recurrent neural networks. In Proceedings of the 20th International Joint Conference on Artificial Intelligence, Hyderabad, India, 6–12 January 2007. [Google Scholar]
- Raghu, M.; Unterthiner, T.; Kornblith, S.; Zhang, C.; Dosovitskiy, A. Do vision transformers see like convolutional neural networks? Adv. Neural Inf. Process. Syst. 2021, 34, 12116–12128. [Google Scholar]
- Maurício, J.; Domingues, I.; Bernardino, J. Comparing vision transformers and convolutional neural networks for image classification: A literature review. Appl. Sci. 2023, 13, 5521. [Google Scholar] [CrossRef]
- Bai, Y.; Mei, J.; Yuille, A.L.; Xie, C. Are transformers more robust than cnns? Adv. Neural Inf. Process. Syst. 2021, 34, 26831–26843. [Google Scholar]
- Tuli, S.; Dasgupta, I.; Grant, E.; Griffiths, T.L. Are convolutional neural networks or transformers more like human vision? arXiv 2021, arXiv:2105.07197. [Google Scholar]
- Ba, J.L.; Kiros, J.R.; Hinton, G.E. Layer normalization. arXiv 2016, arXiv:1607.06450. [Google Scholar]
- Hao, Y.; Dong, L.; Wei, F.; Xu, K. Self-attention attribution: Interpreting information interactions inside transformer. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtual, 2–9 February 2021; Volume 35, pp. 12963–12971. [Google Scholar]
- Liu, Y.; Chen, J.; Chang, Y.; He, S.; Zhou, Z. A novel integration framework for degradation-state prediction via transformer model with autonomous optimizing mechanism. J. Manuf. Syst. 2022, 64, 288–302. [Google Scholar] [CrossRef]
- Casola, S.; Lauriola, I.; Lavelli, A. Pre-trained transformers: An empirical comparison. Mach. Learn. Appl. 2022, 9, 100334. [Google Scholar] [CrossRef]
- Dehghani, M.; Gouws, S.; Vinyals, O.; Uszkoreit, J.; Kaiser, Ł. Universal transformers. arXiv 2018, arXiv:1807.03819. [Google Scholar]
- Raganato, A.; Tiedemann, J. An analysis of encoder representations in transformer-based machine translation. In Proceedings of the 2018 EMNLP Workshop BlackboxNLP: Analyzing and Interpreting Neural Networks for NLP, Brussels, Belgium, 1 November 2018; pp. 287–297. [Google Scholar]
- Wu, K.; Peng, H.; Chen, M.; Fu, J.; Chao, H. Rethinking and improving relative position encoding for vision transformer. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 10033–10041. [Google Scholar]
- Vig, J. A multiscale visualization of attention in the transformer model. arXiv 2019, arXiv:1906.05714. [Google Scholar]
- Xiong, R.; Yang, Y.; He, D.; Zheng, K.; Zheng, S.; Xing, C.; Zhang, H.; Lan, Y.; Wang, L.; Liu, T. On layer normalization in the transformer architecture. In Proceedings of the International Conference on Machine Learning, Virtual, 13–18 July 2020; pp. 10524–10533. [Google Scholar]
- Pan, X.; Ge, C.; Lu, R.; Song, S.; Chen, G.; Huang, Z.; Huang, G. On the integration of self-attention and convolution. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 815–825. [Google Scholar]
- Pu, Q.; Xi, Z.; Yin, S.; Zhao, Z.; Zhao, L. Advantages of transformer and its application for medical image segmentation: A survey. BioMed. Eng. OnLine 2024, 23, 14. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Gheflati, B.; Rivaz, H. Vision transformers for classification of breast ultrasound images. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 480–483. [Google Scholar]
- Zhou, D.; Kang, B.; Jin, X.; Yang, L.; Lian, X.; Jiang, Z.; Hou, Q.; Feng, J. Deepvit: Towards deeper vision transformer. arXiv 2021, arXiv:2103.11886. [Google Scholar]
- Liu, X.; Yu, H.F.; Dhillon, I.; Hsieh, C.J. Learning to encode position for transformer with continuous dynamical model. In Proceedings of the International Conference on Machine Learning, Virtual, 13–18 July 2020; pp. 6327–6335. [Google Scholar]
- Wang, L.; Li, R.; Zhang, C.; Fang, S.; Duan, C.; Meng, X.; Atkinson, P.M. UNetFormer: A UNet-like transformer for efficient semantic segmentation of remote sensing urban scene imagery. ISPRS J. Photogramm. Remote. Sens. 2022, 190, 196–214. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, J.; Shen, C.; Xia, Y. Cotr: Efficiently bridging cnn and transformer for 3d medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021, Proceedings, Part III 24; Springer: Berlin/Heidelberg, Germany, 2021; pp. 171–180. [Google Scholar]
- Zhu, X.; Su, W.; Lu, L.; Li, B.; Wang, X.; Dai, J. Deformable detr: Deformable transformers for end-to-end object detection. arXiv 2020, arXiv:2010.04159. [Google Scholar]
- Yan, X.; Tang, H.; Sun, S.; Ma, H.; Kong, D.; Xie, X. After-unet: Axial fusion transformer unet for medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 3971–3981. [Google Scholar]
- Heidari, M.; Kazerouni, A.; Soltany, M.; Azad, R.; Aghdam, E.K.; Cohen-Adad, J.; Merhof, D. Hiformer: Hierarchical multi-scale representations using transformers for medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 2–7 January 2023; pp. 6202–6212. [Google Scholar]
- Liu, Y.; Wang, H.; Chen, Z.; Huangliang, K.; Zhang, H. TransUNet+: Redesigning the skip connection to enhance features in medical image segmentation. Knowl.-Based Syst. 2022, 256, 109859. [Google Scholar] [CrossRef]
- Chi, J.; Li, Z.; Sun, Z.; Yu, X.; Wang, H. Hybrid transformer UNet for thyroid segmentation from ultrasound scans. Comput. Biol. Med. 2023, 153, 106453. [Google Scholar] [CrossRef]
- Sun, G.; Pan, Y.; Kong, W.; Xu, Z.; Ma, J.; Racharak, T.; Nguyen, L.M.; Xin, J. DA-TransUNet: Integrating spatial and channel dual attention with transformer U-net for medical image segmentation. Front. Bioeng. Biotechnol. 2024, 12, 1398237. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zheng, Y.; Shan, D.; Yang, S.; Li, Q.; Wang, B.; Zhang, Y.; Hong, Q.; Shen, D. Scribformer: Transformer makes cnn work better for scribble-based medical image segmentation. IEEE Trans. Med. Imaging 2024, 43, 2254–2265. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2022; pp. 205–218. [Google Scholar]
- Zhang, J.; Qin, Q.; Ye, Q.; Ruan, T. ST-unet: Swin transformer boosted U-net with cross-layer feature enhancement for medical image segmentation. Comput. Biol. Med. 2023, 153, 106516. [Google Scholar] [CrossRef] [PubMed]
- Azad, R.; Heidari, M.; Shariatnia, M.; Aghdam, E.K.; Karimijafarbigloo, S.; Adeli, E.; Merhof, D. Transdeeplab: Convolution-free transformer-based deeplab v3+ for medical image segmentation. In Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2022; pp. 91–102. [Google Scholar]
- Liang, J.; Yang, C.; Zeng, M.; Wang, X. TransConver: Transformer and convolution parallel network for developing automatic brain tumor segmentation in MRI images. Quant. Imaging Med. Surg. 2022, 12, 2397. [Google Scholar] [CrossRef]
- Yang, H.; Yang, D. CSwin-PNet: A CNN-Swin Transformer combined pyramid network for breast lesion segmentation in ultrasound images. Expert Syst. Appl. 2023, 213, 119024. [Google Scholar] [CrossRef]
- Chen, D.; Yang, W.; Wang, L.; Tan, S.; Lin, J.; Bu, W. PCAT-UNet: UNet-like network fused convolution and transformer for retinal vessel segmentation. PLoS ONE 2022, 17, e0262689. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Chen, Y.; Ji, W.; Yang, F. SSTrans-Net: Smart Swin Transformer Network for medical image segmentation. Biomed. Signal Process. Control. 2024, 91, 106071. [Google Scholar] [CrossRef]
- Pan, S.; Liu, X.; Xie, N.; Chong, Y. EG-TransUNet: A transformer-based U-Net with enhanced and guided models for biomedical image segmentation. BMC Bioinform. 2023, 24, 85. [Google Scholar] [CrossRef] [PubMed]
- Azad, R.; Jia, Y.; Aghdam, E.K.; Cohen-Adad, J.; Merhof, D. Enhancing Medical Image Segmentation with TransCeption: A Multi-Scale Feature Fusion Approach. arXiv 2023, arXiv:2301.10847. [Google Scholar]
- Ma, M.; Xia, H.; Tan, Y.; Li, H.; Song, S. HT-Net: Hierarchical context-attention transformer network for medical ct image segmentation. Appl. Intell. 2022, 52, 10692–10705. [Google Scholar] [CrossRef]
- Huang, S.; Li, J.; Xiao, Y.; Shen, N.; Xu, T. RTNet: Relation transformer network for diabetic retinopathy multi-lesion segmentation. IEEE Trans. Med. Imaging 2022, 41, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Balestra, G.; Zhang, K.; Wang, J.; Rosati, S.; Giannini, V. MultiTrans: Multi-branch transformer network for medical image segmentation. Comput. Methods Programs Biomed. 2024, 254, 108280. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sui, X.; Luo, X.; Xu, X.; Liu, Y.; Goh, R. Medical image segmentation using squeeze-and-expansion transformers. arXiv 2021, arXiv:2105.09511. [Google Scholar]
- Lan, Z.; Chen, M.; Goodman, S.; Gimpel, K.; Sharma, P.; Soricut, R. Albert: A lite bert for self-supervised learning of language representations. arXiv 2019, arXiv:1909.11942. [Google Scholar]
- Chen, P.C.; Tsai, H.; Bhojanapalli, S.; Chung, H.W.; Chang, Y.W.; Ferng, C.S. A simple and effective positional encoding for transformers. arXiv 2021, arXiv:2104.08698. [Google Scholar]
- Shaw, P.; Uszkoreit, J.; Vaswani, A. Self-attention with relative position representations. arXiv 2018, arXiv:1803.02155. [Google Scholar]
- Perera, S.; Navard, P.; Yilmaz, A. SegFormer3D: An Efficient Transformer for 3D Medical Image Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 17–21 June 2024; pp. 4981–4988. [Google Scholar]
- Wang, W.; Chen, C.; Ding, M.; Yu, H.; Zha, S.; Li, J. Transbts: Multimodal brain tumor segmentation using transformer. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021, Proceedings, Part I 24; Springer: Berlin/Heidelberg, Germany, 2021; pp. 109–119. [Google Scholar]
- Cuenat, S.; Couturier, R. Convolutional neural network (cnn) vs. vision transformer (vit) for digital holography. In Proceedings of the 2022 2nd International Conference on Computer, Control and Robotics (ICCCR), Shanghai, China, 18–20 March 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 235–240. [Google Scholar]
- Zhou, H.Y.; Lu, C.; Yang, S.; Yu, Y. Convnets vs. transformers: Whose visual representations are more transferable? In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 2230–2238. [Google Scholar]
- Tang, Y.; Yang, D.; Li, W.; Roth, H.R.; Landman, B.; Xu, D.; Nath, V.; Hatamizadeh, A. Self-supervised pre-training of swin transformers for 3d medical image analysis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 20730–20740. [Google Scholar]
- You, C.; Zhao, R.; Staib, L.H.; Duncan, J.S. Momentum contrastive voxel-wise representation learning for semi-supervised volumetric medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 639–652. [Google Scholar]
- You, C.; Dai, W.; Min, Y.; Staib, L.; Duncan, J.S. Implicit anatomical rendering for medical image segmentation with stochastic experts. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2023; Springer: Berlin/Heidelberg, Germany, 2023; pp. 561–571. [Google Scholar]
- Zhu, X.; Goldberg, A.B. Introduction to Semi-Supervised Learning; Springer Nature: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
- Jiang, J.; Veeraraghavan, H. Self-Supervised Pretraining in the Wild Imparts Image Acquisition Robustness to Medical Image Transformers: An Application to Lung Cancer Segmentation. Medical Imaging with Deep Learning. 2024. Available online: https://openreview.net/forum?id=G9Te2IevNm (accessed on 1 July 2024).
- Cai, Z.; Ravichandran, A.; Favaro, P.; Wang, M.; Modolo, D.; Bhotika, R.; Tu, Z.; Soatto, S. Semi-supervised vision transformers at scale. Adv. Neural Inf. Process. Syst. 2022, 35, 25697–25710. [Google Scholar]
- You, C.; Zhao, R.; Liu, F.; Dong, S.; Chinchali, S.; Topcu, U.; Staib, L.; Duncan, J. Class-aware adversarial transformers for medical image segmentation. Adv. Neural Inf. Process. Syst. 2022, 35, 29582–29596. [Google Scholar]
- Arkin, E.; Yadikar, N.; Xu, X.; Aysa, A.; Ubul, K. A survey: Object detection methods from CNN to transformer. Multimed. Tools Appl. 2023, 82, 21353–21383. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, T.; Yu, F.; Dai, J.; Konukoglu, E.; Van Gool, L. Exploring cross-image pixel contrast for semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 7303–7313. [Google Scholar]
- You, C.; Dai, W.; Min, Y.; Liu, F.; Clifton, D.; Zhou, S.K.; Staib, L.; Duncan, J. Rethinking semi-supervised medical image segmentation: A variance-reduction perspective. Adv. Neural Inf. Process. Syst. 2024, 36, 9984–10021. [Google Scholar]
- Xu, Z.; Dai, Y.; Liu, F.; Wu, B.; Chen, W.; Shi, L. Swin MoCo: Improving parotid gland MRI segmentation using contrastive learning. Med. Phys. 2024. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Xie, S.; Lin, L.; Tong, R.; Chen, Y.W.; Wang, H.; Li, Y.; Huang, Y.; Zheng, Y. ClassFormer: Exploring class-aware dependency with transformer for medical image segmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, Washington DC, USA, 7–14 February 2023; Volume 37, pp. 917–925. [Google Scholar]
- Yuan, N.; Zhang, Y.; Lv, K.; Liu, Y.; Yang, A.; Hu, P.; Yu, H.; Han, X.; Guo, X.; Li, J.; et al. HCA-DAN: Hierarchical class-aware domain adaptive network for gastric tumor segmentation in 3D CT images. Cancer Imaging 2024, 24, 63. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Lin, X.; Yang, X.; Yu, L.; Cheng, K.T.; Yan, Z. UCTNet: Uncertainty-guided CNN-Transformer hybrid networks for medical image segmentation. Pattern Recognit. 2024, 152, 110491. [Google Scholar] [CrossRef]
- Xiao, Z.; Sun, H.; Liu, F. Semi-supervised CT image segmentation via contrastive learning based on entropy constraints. Biomed. Eng. Lett. 2024, 1–13. [Google Scholar] [CrossRef]
- Wu, Y.; Li, X.; Zhou, Y. Uncertainty-aware representation calibration for semi-supervised medical imaging segmentation. Neurocomputing 2024, 595, 127912. [Google Scholar] [CrossRef]
- Zhao, X.; Qi, Z.; Wang, S.; Wang, Q.; Wu, X.; Mao, Y.; Zhang, L. Rcps: Rectified contrastive pseudo supervision for semi-supervised medical image segmentation. IEEE J. Biomed. Health Inform. 2023, 28, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Azad, R.; Aghdam, E.K.; Rauland, A.; Jia, Y.; Avval, A.H.; Bozorgpour, A.; Karimijafarbigloo, S.; Cohen, J.P.; Adeli, E.; Merhof, D. Medical image segmentation review: The success of u-net. arXiv 2022, arXiv:2211.14830. [Google Scholar]
- He, K.; Gan, C.; Li, Z.; Rekik, I.; Yin, Z.; Ji, W.; Gao, Y.; Wang, Q.; Zhang, J.; Shen, D. Transformers in medical image analysis. Intell. Med. 2022, 3, 59–78. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Mehrani, P.; Tsotsos, J.K. Self-attention in vision transformers performs perceptual grouping, not attention. arXiv 2023, arXiv:2303.01542. [Google Scholar] [CrossRef]
- Han, X.; Zhang, Z.; Ding, N.; Gu, Y.; Liu, X.; Huo, Y.; Qiu, J.; Yao, Y.; Zhang, A.; Zhang, L.; et al. Pre-trained models: Past, present and future. AI Open 2021, 2, 225–250. [Google Scholar] [CrossRef]
- Radford, A.; Narasimhan, K.; Salimans, T.; Sutskever, I. Improving Language Understanding by Generative Pre-Training. 2018. Available online: https://www.mikecaptain.com/resources/pdf/GPT-1.pdf (accessed on 1 July 2024).
- Team, G.; Anil, R.; Borgeaud, S.; Wu, Y.; Alayrac, J.B.; Yu, J.; Soricut, R.; Schalkwyk, J.; Dai, A.M.; Hauth, A.; et al. Gemini: A family of highly capable multimodal models. arXiv 2023, arXiv:2312.11805. [Google Scholar]
- Du, G.; Cao, X.; Liang, J.; Chen, X.; Zhan, Y. Medical image segmentation based on u-net: A review. J. Imaging Sci. Technol. 2020, 64, 020508-1–020508-12. [Google Scholar] [CrossRef]
- Chen, H.; Dong, Y.; Lu, Z.; Yu, Y.; Han, J. Pixel Matching Network for Cross-Domain Few-Shot Segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2024; pp. 978–987. [Google Scholar]
- Li, Z.; Chen, Z.; Liu, X.; Jiang, J. Depthformer: Exploiting long-range correlation and local information for accurate monocular depth estimation. Mach. Intell. Res. 2023, 20, 837–854. [Google Scholar] [CrossRef]




| Characteristics | Self-Attention Mechanism | Convolutional Operation |
|---|---|---|
| Applicability | Suitable for long-range dependencies. | Suitable for extracting local features and structures. |
| Fully connected; each element can influence all others. | Locally connected; each neuron relates to a small portion of the input. | |
| Parameter Count | More parameters; requires more computational resources. | Fewer parameters; more computationally efficient. |
| Computational Efficiency | Higher computational complexity. | Lower computational complexity, particularly for large-scale data. |
| Translation Invariance | Lacks translation invariance; sensitive to position. | Possesses translation invariance; insensitive to position. |
| Often requires position encoding for handling sequence information. | No need for additional position encoding. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Deng, X.; Ling, S.H. Next-Gen Medical Imaging: U-Net Evolution and the Rise of Transformers. Sensors 2024, 24, 4668. https://doi.org/10.3390/s24144668
Zhang C, Deng X, Ling SH. Next-Gen Medical Imaging: U-Net Evolution and the Rise of Transformers. Sensors. 2024; 24(14):4668. https://doi.org/10.3390/s24144668
Chicago/Turabian StyleZhang, Chen, Xiangyao Deng, and Sai Ho Ling. 2024. "Next-Gen Medical Imaging: U-Net Evolution and the Rise of Transformers" Sensors 24, no. 14: 4668. https://doi.org/10.3390/s24144668
APA StyleZhang, C., Deng, X., & Ling, S. H. (2024). Next-Gen Medical Imaging: U-Net Evolution and the Rise of Transformers. Sensors, 24(14), 4668. https://doi.org/10.3390/s24144668







