An Unsupervised Learning Tool for Plaque Tissue Characterization in Histopathological Images
Abstract
1. Introduction
2. Material and Methods
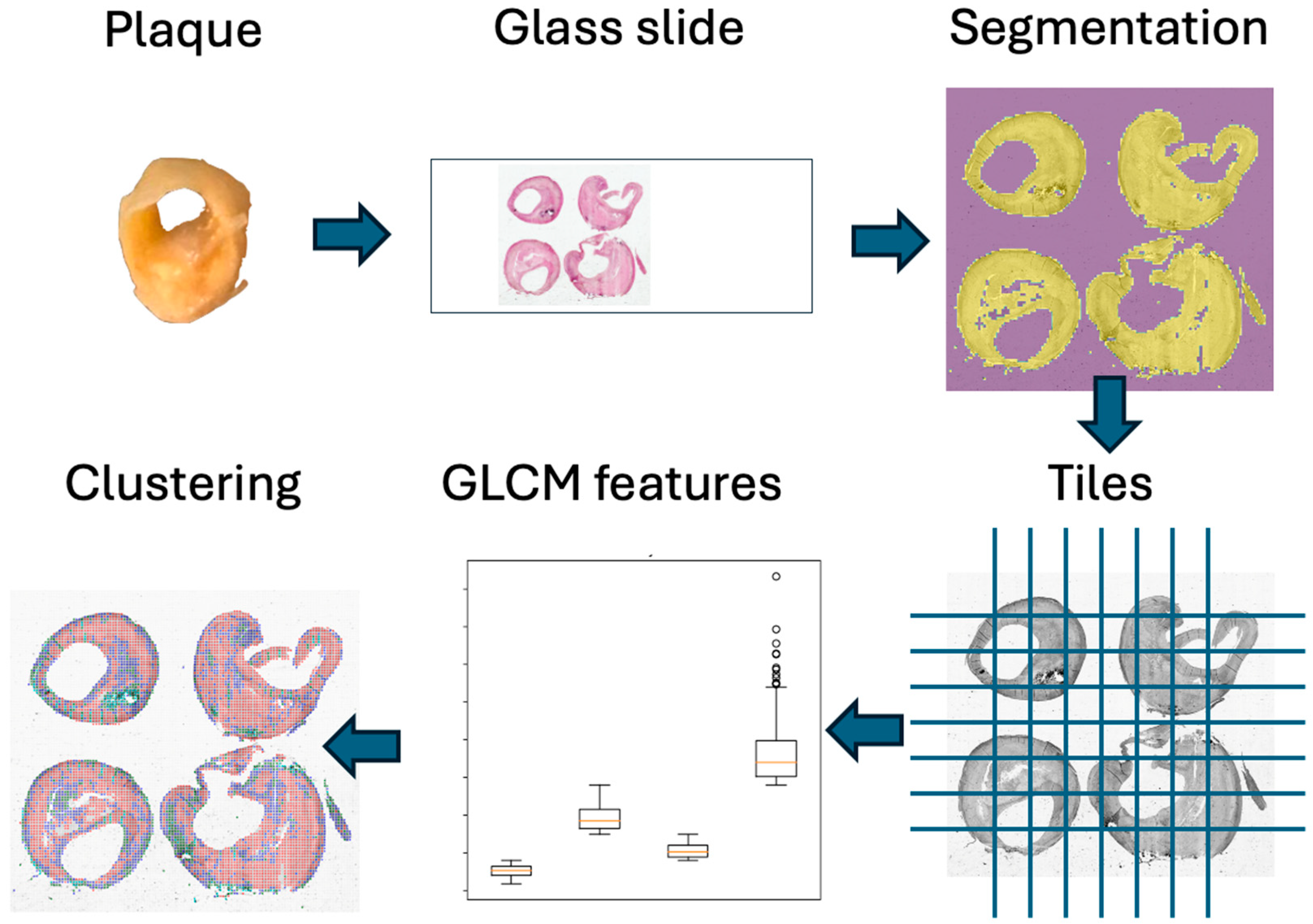
2.1. Image Acquisition
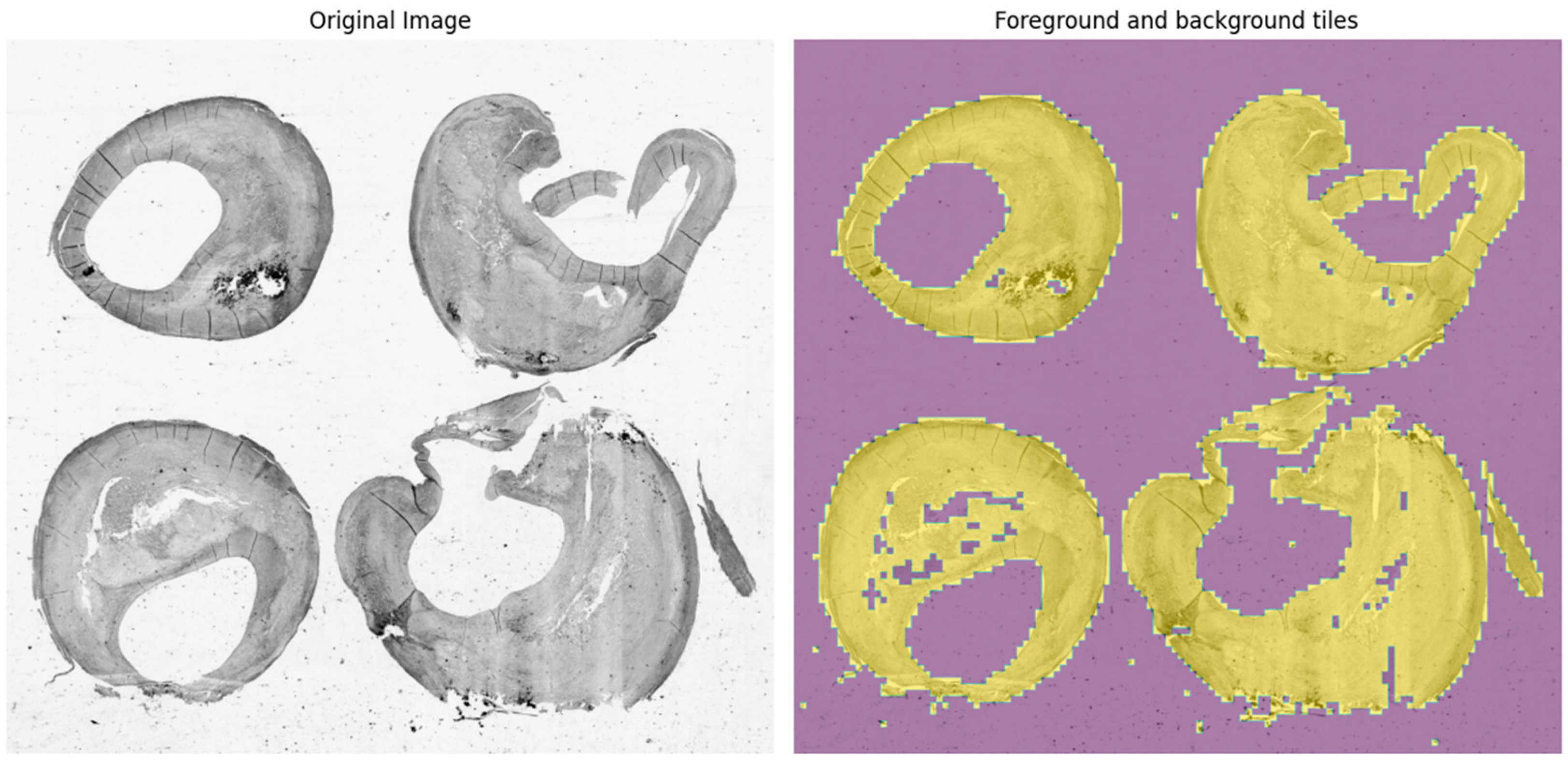
2.2. Image Processing
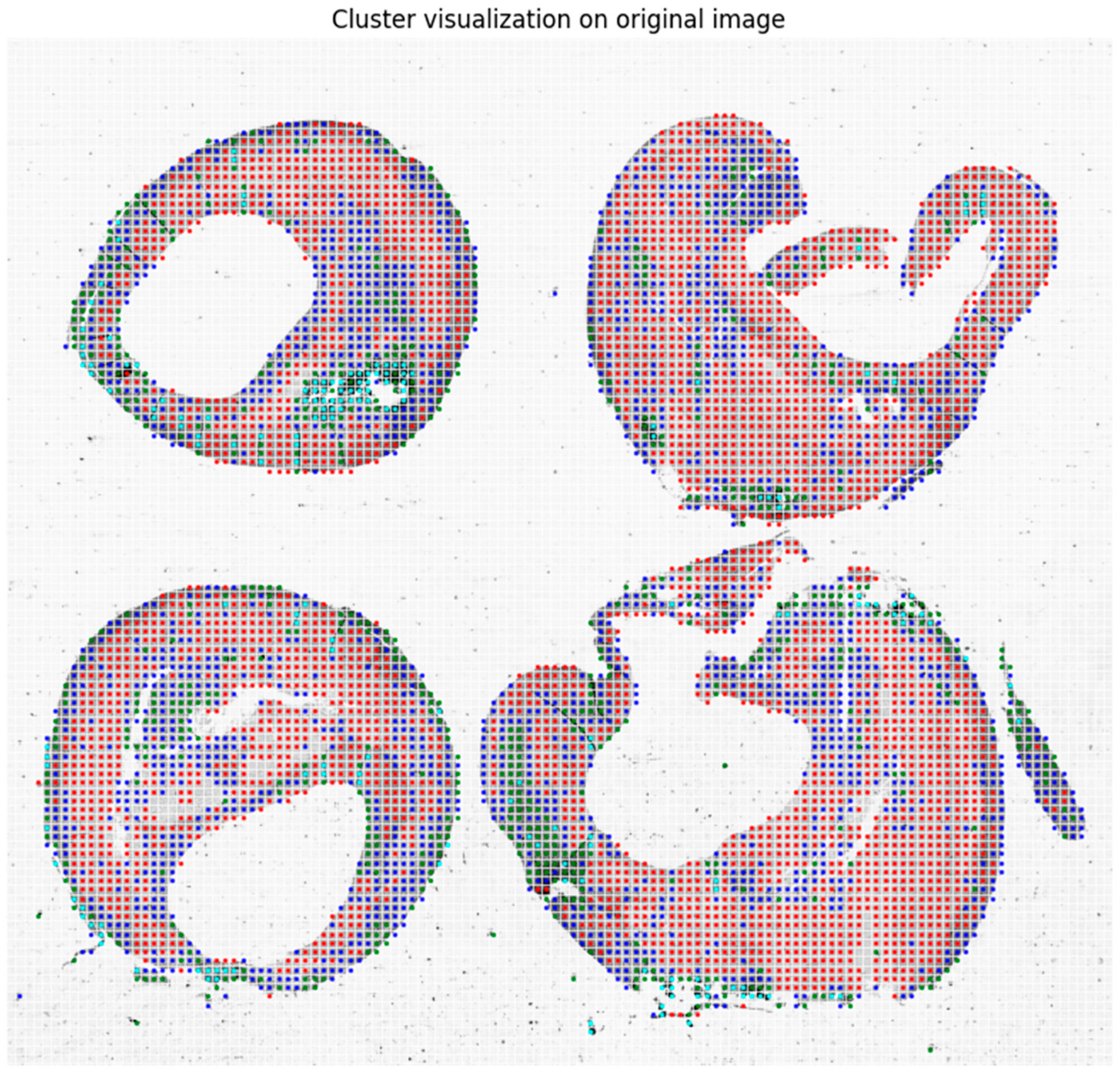
2.3. Clustering
2.4. Unsupervised Evaluation
2.5. Replication
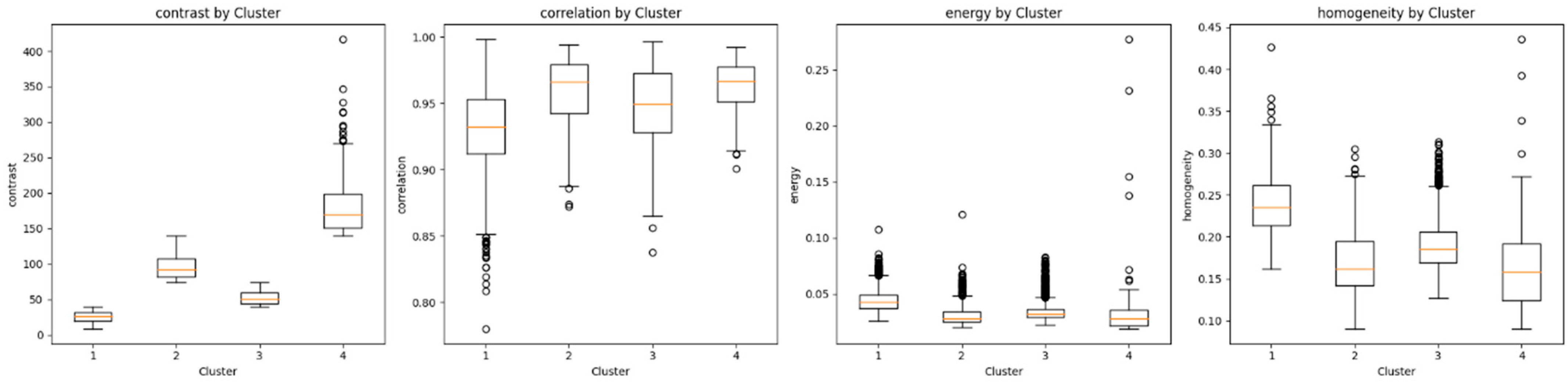
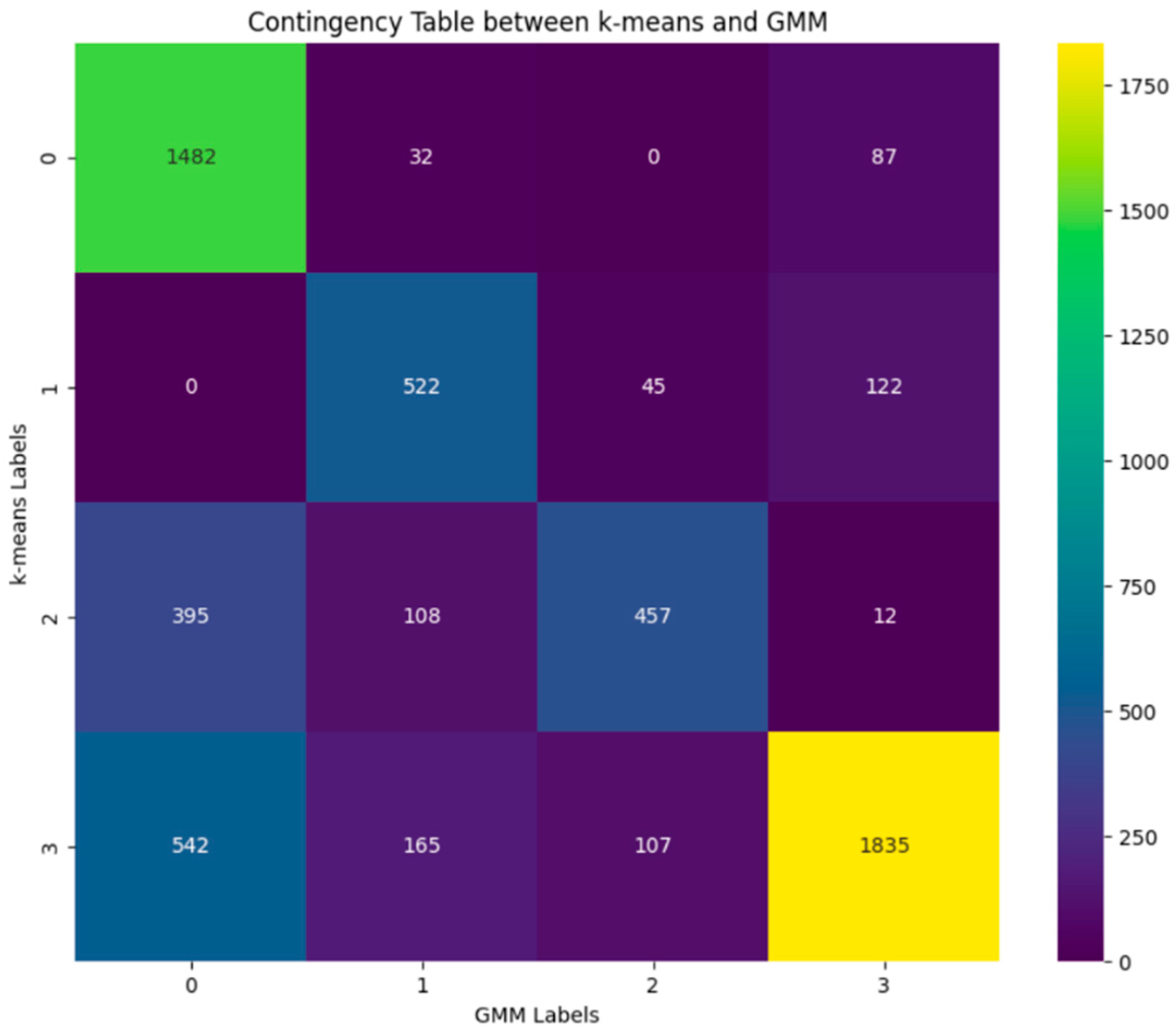
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Katan, M.; Luft, A. Global Burden of Stroke. Semin. Neurol. 2018, 38, 208–211. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Aravkin, A.Y.; Zheng, P.; Abbafati, C.; Abbas, K.M.; Abbasi-Kangevari, M.; Abd-Allah, F.; Abdelalim, A.; Abdollahi, M.; Abdollahpour, I.; et al. Global Burden of 87 Risk Factors in 204 Countries and Territories, 1990–2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1223–1249. [Google Scholar] [CrossRef]
- Fan, J.; Li, X.; Yu, X.; Liu, Z.; Jiang, Y.; Fang, Y.; Zong, M.; Suo, C.; Man, Q.; Xiong, L. Global Burden, Risk Factor Analysis, and Prediction Study of Ischemic Stroke, 1990–2030. Neurology 2023, 101, e137–e150. [Google Scholar] [CrossRef]
- Pu, L.; Wang, L.; Zhang, R.; Zhao, T.; Jiang, Y.; Han, L. Projected Global Trends in Ischemic Stroke Incidence, Deaths and Disability-Adjusted Life Years From 2020 to 2030. Stroke 2023, 54, 1330–1339. [Google Scholar] [CrossRef]
- Yahagi, K.; Kolodgie, F.D.; Lutter, C.; Mori, H.; Romero, M.E.; Finn, A.V.; Virmani, R. Pathology of Human Coronary and Carotid Artery Atherosclerosis and Vascular Calcification in Diabetes Mellitus. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Munger, E.; Hickey, J.W.; Dey, A.K.; Jafri, M.S.; Kinser, J.M.; Mehta, N.N. Application of Machine Learning in Understanding Atherosclerosis: Emerging Insights. APL Bioeng. 2021, 5, 011505. [Google Scholar] [CrossRef]
- Kamtchum-Tatuene, J.; Saba, L.; Heldner, M.R.; Poorthuis, M.H.F.; de Borst, G.J.; Rundek, T.; Kakkos, S.K.; Chaturvedi, S.; Topakian, R.; Polak, J.F.; et al. Interleukin-6 Predicts Carotid Plaque Severity, Vulnerability, and Progression. Circ. Res. 2022, 131, e22–e33. [Google Scholar] [CrossRef]
- Goncalves, I.; Sun, J.; Tengryd, C.; Nitulescu, M.; Persson, A.F.; Nilsson, J.; Edsfeldt, A. Plaque Vulnerability Index Predicts Cardiovascular Events: A Histological Study of an Endarterectomy Cohort. J. Am. Heart Assoc. 2021, 10, e021038. [Google Scholar] [CrossRef] [PubMed]
- Bentzon, J.F.; Otsuka, F.; Virmani, R.; Falk, E. Mechanisms of Plaque Formation and Rupture. Circ. Res. 2014, 114, 1852–1866. [Google Scholar] [CrossRef] [PubMed]
- Libby, P. The Changing Landscape of Atherosclerosis. Nature 2021, 592, 524–533. [Google Scholar] [CrossRef]
- Saba, L.; Saam, T.; Jäger, H.R.; Yuan, C.; Hatsukami, T.S.; Saloner, D.; Wasserman, B.A.; Bonati, L.H.; Wintermark, M. Imaging Biomarkers of Vulnerable Carotid Plaques for Stroke Risk Prediction and Their Potential Clinical Implications. Lancet Neurol. 2019, 18, 559–572. [Google Scholar] [CrossRef]
- Stary, H.C.; Chandler, A.B.; Dinsmore, R.E.; Fuster, V.; Glagov, S.; Insull, W.; Rosenfeld, M.E.; Schwartz, C.J.; Wagner, W.D.; Wissler, R.W. A Definition of Advanced Types of Atherosclerotic Lesions and a Histological Classification of Atherosclerosis. A Report from the Committee on Vascular Lesions of the Council on Arteriosclerosis, American Heart Association. Circulation 1995, 92, 1355–1374. [Google Scholar] [CrossRef]
- Faa, G.; Cau, R.; Ravarino, A.; Canino, A.; Van Eyken, P.; Fraschini, M.; Suri, J.S.; Saba, L. Lessons from Autopsy: Topographical Variability of Atherosclerosis Plaques. 2024. Available online: https://journals.sagepub.com/doi/full/10.1177/22799036241249659 (accessed on 30 May 2024).
- Sirimarco, G.; Amarenco, P.; Labreuche, J.; Touboul, P.-J.; Alberts, M.; Goto, S.; Rother, J.; Mas, J.-L.; Bhatt, D.L.; Steg, P.G.; et al. Carotid Atherosclerosis and Risk of Subsequent Coronary Event in Outpatients with Atherothrombosis. Stroke 2013, 44, 373–379. [Google Scholar] [CrossRef]
- Bytyçi, I.; Shenouda, R.; Wester, P.; Henein, M.Y. Carotid Atherosclerosis in Predicting Coronary Artery Disease: A Systematic Review and Meta-Analysis. Arter. Thromb. Vasc. Biol. 2021, 41, e224–e237. [Google Scholar] [CrossRef]
- Hertzer, N.R.; Young, J.R.; Beven, E.G.; Graor, R.A.; O’Hara, P.J.; Ruschhaupt, W.F.; de Wolfe, V.G.; Maljovec, L.C. Coronary Angiography in 506 Patients with Extracranial Cerebrovascular Disease. Arch. Intern. Med. 1985, 145, 849–852. [Google Scholar] [CrossRef]
- Faa, G.; Castagnola, M.; Didaci, L.; Coghe, F.; Scartozzi, M.; Saba, L.; Fraschini, M. The Quest for the Application of Artificial Intelligence to Whole Slide Imaging: Unique Prospective from New Advanced Tools. Algorithms 2024, 17, 254. [Google Scholar] [CrossRef]
- Ariotta, V.; Lehtonen, O.; Salloum, S.; Micoli, G.; Lavikka, K.; Rantanen, V.; Hynninen, J.; Virtanen, A.; Hautaniemi, S. H&E Image Analysis Pipeline for Quantifying Morphological Features. J. Pathol. Inform. 2023, 14, 100339. [Google Scholar] [CrossRef]
- Al-Thelaya, K.; Gilal, N.U.; Alzubaidi, M.; Majeed, F.; Agus, M.; Schneider, J.; Househ, M. Applications of Discriminative and Deep Learning Feature Extraction Methods for Whole Slide Image Analysis: A Survey. J. Pathol. Inform. 2023, 14, 100335. [Google Scholar] [CrossRef]
- Jenkinson, E.; Arandjelović, O. Whole Slide Image Understanding in Pathology: What Is the Salient Scale of Analysis? BioMedInformatics 2024, 4, 489–518. [Google Scholar] [CrossRef]
- van der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T. Scikit-Image: Image Processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Sezgin, M.; Sankur, B. Survey over Image Thresholding Techniques and Quantitative Performance Evaluation. J. Electron. Imaging 2004, 13, 146–165. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A K-Means Clustering Algorithm. J. R. Stat. Soc. Ser. C (Appl. Stat.) 1979, 28, 100–108. [Google Scholar] [CrossRef]
- MacQueen, J. Some Methods for Classification and Analysis of Multivariate Observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Volume 1: Statistics; University of California Press: Berkeley, CA, USA, 1967; Volume 5.1, pp. 281–298. [Google Scholar]
- Thorndike, R.L. Who Belongs in the Family? Psychometrika 1953, 18, 267–276. [Google Scholar] [CrossRef]
- Schubert, E. Stop Using the Elbow Criterion for K-Means and How to Choose the Number of Clusters Instead. SIGKDD Explor. Newsl. 2023, 25, 36–42. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A Graphical Aid to the Interpretation and Validation of Cluster Analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Monshizadeh, M.; Khatri, V.; Kantola, R.; Yan, Z. A Deep Density Based and Self-Determining Clustering Approach to Label Unknown Traffic. J. Netw. Comput. Appl. 2022, 207, 103513. [Google Scholar] [CrossRef]
- Davies, D.L.; Bouldin, D.W. A Cluster Separation Measure. IEEE Trans. Pattern Anal. Mach. Intell. 1979, PAMI-1, 224–227. [Google Scholar] [CrossRef]
- Patel, E.; Kushwaha, D.S. Clustering Cloud Workloads: K-Means vs. Gaussian Mixture Model. Procedia Comput. Sci. 2020, 171, 158–167. [Google Scholar] [CrossRef]
- Berkhin, P. A Survey of Clustering Data Mining Techniques. In Grouping Multidimensional Data: Recent Advances in Clustering; Kogan, J., Nicholas, C., Teboulle, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 25–71. ISBN 978-3-540-28349-2. [Google Scholar]
- Bishop, C.M.; Nasrabadi, N.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4. [Google Scholar]
- Miceli, G.; Rizzo, G.; Basso, M.G.; Cocciola, E.; Pennacchio, A.R.; Pintus, C.; Tuttolomondo, A. Artificial Intelligence in Symptomatic Carotid Plaque Detection: A Narrative Review. Appl. Sci. 2023, 13, 4321. [Google Scholar] [CrossRef]
- Cau, R.; Flanders, A.; Mannelli, L.; Politi, C.; Faa, G.; Suri, J.S.; Saba, L. Artificial Intelligence in Computed Tomography Plaque Characterization: A Review. Eur. J. Radiol. 2021, 140, 109767. [Google Scholar] [CrossRef]
- Li, R.; Zheng, J.; Zayed, M.A.; Saffitz, J.E.; Woodard, P.K.; Jha, A.K. Carotid Atherosclerotic Plaque Segmentation in Multi-Weighted MRI Using a Two-Stage Neural Network: Advantages of Training with High-Resolution Imaging and Histology. Front. Cardiovasc. Med. 2023, 10, 1127653. [Google Scholar] [CrossRef]
- Saba, L.; Sanagala, S.S.; Gupta, S.K.; Koppula, V.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M.; et al. Multimodality Carotid Plaque Tissue Characterization and Classification in the Artificial Intelligence Paradigm: A Narrative Review for Stroke Application. Ann. Transl. Med. 2021, 9, 1206. [Google Scholar] [CrossRef]
- Chen, J.; Wang, S.; Wang, K.; Abiri, P.; Huang, Z.; Yin, J.; Jabalera, A.M.; Arianpour, B.; Roustaei, M.; Zhu, E.; et al. Machine Learning-directed Electrical Impedance Tomography to Predict Metabolically Vulnerable Plaques. Bioeng. Transl. Med. 2023, 9, e10616. [Google Scholar] [CrossRef]
- Caballero, R.; Martínez, M.Á.; Peña, E. Coronary Artery Properties in Atherosclerosis: A Deep Learning Predictive Model. Front. Physiol. 2023, 14, 1162436. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Li, Z.; Wang, J.; Huang, Y.; Yin, Y.; Li, Z. Atherosclerotic Plaque Tissue Characterization: An OCT-Based Machine Learning Algorithm With Ex Vivo Validation. Front. Bioeng. Biotechnol. 2020, 8, 749. [Google Scholar] [CrossRef]
- Cooper, M.; Ji, Z.; Krishnan, R.G. Machine Learning in Computational Histopathology: Challenges and Opportunities. Genes Chromosomes Cancer 2023, 62, 540–556. [Google Scholar] [CrossRef]
- Berman, A.G.; Orchard, W.R.; Gehrung, M.; Markowetz, F. SliDL: A Toolbox for Processing Whole-Slide Images in Deep Learning. PLoS ONE 2023, 18, e0289499. [Google Scholar] [CrossRef]
- Lee, M. Recent Advancements in Deep Learning Using Whole Slide Imaging for Cancer Prognosis. Bioengineering 2023, 10, 897. [Google Scholar] [CrossRef] [PubMed]
- Naik, D.A.; Mohana, R.M.; Ramu, G.; Lalitha, Y.S.; SureshKumar, M.; Raghavender, K.V. Analyzing Histopathological Images by Using Machine Learning Techniques. Appl. Nanosci. 2023, 13, 2507–2513. [Google Scholar] [CrossRef]
- Chen, C.; Lu, M.Y.; Williamson, D.F.K.; Chen, T.Y.; Schaumberg, A.J.; Mahmood, F. Fast and Scalable Search of Whole-Slide Images via Self-Supervised Deep Learning. Nat. Biomed. Eng. 2022, 6, 1420–1434. [Google Scholar] [CrossRef] [PubMed]
- Song, A.H.; Jaume, G.; Williamson, D.F.K.; Lu, M.Y.; Vaidya, A.; Miller, T.R.; Mahmood, F. Artificial Intelligence for Digital and Computational Pathology. Nat. Rev. Bioeng. 2023, 1, 930–949. [Google Scholar] [CrossRef]
- van der Laak, J.; Litjens, G.; Ciompi, F. Deep Learning in Histopathology: The Path to the Clinic. Nat. Med. 2021, 27, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Amato, D.; Calderaro, S.; Lo Bosco, G.; Rizzo, R.; Vella, F. Metric Learning in Histopathological Image Classification: Opening the Black Box. Sensors 2023, 23, 6003. [Google Scholar] [CrossRef]
- Vulli, A.; Srinivasu, P.N.; Sashank, M.S.K.; Shafi, J.; Choi, J.; Ijaz, M.F. Fine-Tuned DenseNet-169 for Breast Cancer Metastasis Prediction Using FastAI and 1-Cycle Policy. Sensors 2022, 22, 2988. [Google Scholar] [CrossRef]
- Song, Y.; Cisternino, F.; Mekke, J.M.; de Borst, G.J.; de Kleijn, D.P.V.; Pasterkamp, G.; Vink, A.; Glastonbury, C.A.; van der Laan, S.W.; Miller, C.L. An Automatic Entropy Method to Efficiently Mask Histology Whole-Slide Images. Sci. Rep. 2023, 13, 4321. [Google Scholar] [CrossRef]



| Size | K | Silhouette Score | Davies-Bouldin Index | ARI | NMI |
|---|---|---|---|---|---|
| 30 × 30 | 4 | 0.617 | 0.52 | 0.362 | 0.398 |
| 50 × 50 | 4 | 0.581 | 0.54 | 0.405 | 0.412 |
| 75 × 75 | 4 | 0.569 | 0.55 | 0.273 | 0.362 |
| 100 × 100 | 4 | 0.566 | 0.55 | 0.438 | 0.448 |
| 150 × 150 | 4 | 0.545 | 0.52 | 0.264 | 0.359 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fraschini, M.; Castagnola, M.; Barberini, L.; Sanfilippo, R.; Coghe, F.; Didaci, L.; Cau, R.; Frongia, C.; Scartozzi, M.; Saba, L.; et al. An Unsupervised Learning Tool for Plaque Tissue Characterization in Histopathological Images. Sensors 2024, 24, 5383. https://doi.org/10.3390/s24165383
Fraschini M, Castagnola M, Barberini L, Sanfilippo R, Coghe F, Didaci L, Cau R, Frongia C, Scartozzi M, Saba L, et al. An Unsupervised Learning Tool for Plaque Tissue Characterization in Histopathological Images. Sensors. 2024; 24(16):5383. https://doi.org/10.3390/s24165383
Chicago/Turabian StyleFraschini, Matteo, Massimo Castagnola, Luigi Barberini, Roberto Sanfilippo, Ferdinando Coghe, Luca Didaci, Riccardo Cau, Claudio Frongia, Mario Scartozzi, Luca Saba, and et al. 2024. "An Unsupervised Learning Tool for Plaque Tissue Characterization in Histopathological Images" Sensors 24, no. 16: 5383. https://doi.org/10.3390/s24165383
APA StyleFraschini, M., Castagnola, M., Barberini, L., Sanfilippo, R., Coghe, F., Didaci, L., Cau, R., Frongia, C., Scartozzi, M., Saba, L., & Faa, G. (2024). An Unsupervised Learning Tool for Plaque Tissue Characterization in Histopathological Images. Sensors, 24(16), 5383. https://doi.org/10.3390/s24165383








