1. Introduction
Cardiovascular diseases pose long-term risks to human health and the trend of cardiovascular diseases affecting younger populations has become increasingly prominent in recent years [
1]. Therefore, early and effective diagnosis of heart disease is extremely important. The heart possesses multimodal characteristics [
2]. During an exercise cycle, the cardiac sinus node sends out electrical signals that are transmitted to the cardiomyocytes to produce contraction and diastole of the myocardium, which drives the blood to flow. This process generates various mechanical wave vibrations. Ballistocardiography (BCG, whole-body motion produced by blood ejection into a blood vessel) [
3] and seismocardiography (SCG, localized vibration of the chest wall induced by a heartbeat) [
4] are able to represent the process of myocardial systolic-diastolic motion. Phonocardiography (PCG, 20–100 Hz) [
5] is a sound signal generated by blood flow during the opening and closing of valves. Gyrocardiography (GCG) [
6] embodies information about the cardiac rotational motion in different directions. The vibrational waves described above are essential for people to recognize information about cardiac motion, reflecting structural abnormalities and altered ejection capacity of the heart. The 24-h ECG Holter reflects the daily electrical behavior of the heart but the ECG signal alone does not reflect the mechanical information described above. The realization of dynamic monitoring of cardiac mechanical vibration can expand the dimensions of cardiac monitoring [
7] and the combination of electrical signals can reflect more comprehensive cardiac information.
Since the 1990s, miniaturized and lightweight mechanical vibration sensors have been increasingly utilized in portable cardiac monitoring systems. Accelerometers based on microelectromechanical systems (MEMS) technology are widely used for SCG detection [
8,
9] and have been extended to a variety of application scenarios, including heart rate detection [
10], assessment of hemodynamic parameters such as blood pressure [
11] and myocardial contractility [
12], and postoperative patient monitoring [
13]. MEMS-based gyroscopes are used to extract GCG [
6]. An Inertial Measurement Unit (IMU) allows the simultaneous extraction of 3-axis SCG signals and 3-axis GCG signals on a single miniaturized device, obtaining more information than a single sensing method [
14]. A wearable hermetically sealed high-precision vibration sensor combines the properties of an accelerometer and a contact microphone to monitor a wide range of health information induced by cardiopulmonary vibration [
15]. A stretchable sensor incorporating soft gold electrodes can be attached directly to the skin to extract cardiac time intervals [
11]. Deformable conductive bioelectronics are increasingly being used to monitor the electrophysiologic state of the heart to improve diagnostic accuracy [
16].
Based on the extraction of various vibration waves, people have gradually established a macroscopic connection between them. There is an overlap of frequencies between the vibration waves, so they are correlated. For example, both SCG (energy concentrated at 0.5–30 Hz) and PCG (energy concentrated at 20–200 Hz) can extract information about valve closure (MC, AO, AC, and MO for SCG as well as S1 and S2 for PCG). Researchers identified the vibration corresponding to PCG using vibrocardiography (SCG and GCG) [
17], extracted the PCG process from SCG [
18], and annotated the features of SCG by GCG [
19]. Meanwhile, the vibration wave reflects different motion information of the heart because of the various frequencies. Among the heart signals extracted from the accelerometer, the band-pass filtered signal of 18–200 Hz represents the information of opening and closing of the heart valves, while the band-pass filtered signal of 0.6–20 Hz embodies the information of the recoil force of the heart contraction [
20]. In more detail, signals below 0.5 Hz characterize respiratory rhythms [
21]; signals around 1–5 Hz characterize cardiac volume changes and ejection phase activity [
22,
23]; SCG signals around 5–30 Hz characterize the opening and closing of cardiac valves as well as the systolic and diastolic processes [
22]; and PCG signals around 20–200 Hz characterize high-frequency acoustic activity. Recently, researchers have been using piezoelectric transducers to synchronously extract signals in multiple frequency bands as described above [
22,
23,
24], thanks to the broadband nature of piezoelectric methods. Simultaneous passive sensing is another advantage of piezoelectric methods, which can generate charges without an external power source. Therefore, a miniaturized high-sensitive piezoelectric transducer module can be designed for multi-frequency vibration detection in the heart, allowing the extraction of comprehensive mechanical information about the heart.
Furthermore, dynamic monitoring of cardiac activity relies on accurate and efficient feature extraction methods and researchers are currently keen to investigate automatic annotation algorithms. For ULF-SCG below 5 Hz, the ‘findpeaks’ function (from MATLAB) can extract the locations of the feature points (peaks and troughs). For PCG above 200 Hz, the locations of S1 and S2 can be identified using the energy spectrum and envelope methods [
25]. The SCG signals around 5–30 Hz cover the infrasound and audible bands and have nine feature points. Because of the weak interpretability and high variability of waveform morphology, the feature extraction of SCG has become a difficult task in research. Currently, most methods can only extract some feature points with high-frequency bands. The Gaussian mixture model approach extracted two feature points (AC and AO) with low latency [
26]; researchers achieved the labeling of IM and AC of SCGs by using the envelopes of the high-frequency signals from the same accelerometer [
27]; and the wavelet transform approach can locate both AO and IM points [
28]. The binary classifier approach is capable of extracting multiple feature points but the introduction of integrated features makes the results dependent on the morphology of the waveform [
29]; the curvature method [
22] is capable of extracting multiple feature points regardless of the waveform morphology but sacrifices timeliness due to its susceptibility to interference and the need for single-cycle averaging processing to achieve high recognition accuracy. Therefore, it becomes a challenge to meet the requirements of high recognition of morphological variability robustness, low latency, and handling multiple feature points.
Recently, 1D-CNN approaches have become attractive for complex engineering because they do not require manual feature production and can directly extract “learned features” [
30]. Some researchers have performed 1D to 2D conversion to apply deep CNN methods [
31] but the high computational complexity makes them unsuitable for real-time operation on mobile and low-power/low-memory devices. Compact adaptive 1D-CNN can [
32] operate directly on one-dimensional physiological signals with low time complexity. Early arrhythmia detection in ECG beats [
33] and feature extraction of pulse waves [
34] are successful 1D-CNN applications.
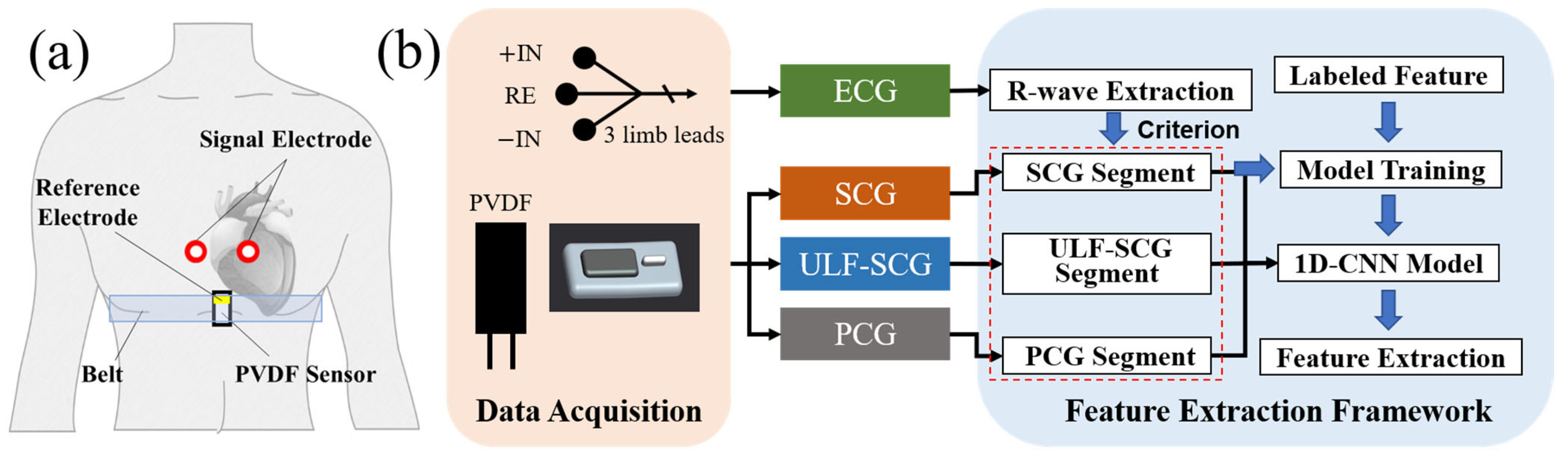
In this study, first of all, we described the multi-frequency vibration model of the heart. Combined with Fourier series theory, the heart vibration is represented as a synthesis of vibration waves in different frequency bands and the signal is validated using EMD decomposition. Then, we propose a sensor module based on the PVDF piezoelectric film for simultaneous acquisition of multi-frequency vibrations of the heart allowing detecting both ultra-low frequency cardiac signals (ULF-SCG) and SCG signals [
22]. Besides, we further optimized the volume of the sensor and designed a separate structure. Comparative experiments verified that the sensor can also extract PCG signals simultaneously. The new sensor module achieves an output sensitivity of 40.6 V/N for the module with a mass of 2.4 g in a volume of 30 × 15 × 5 mm. Meanwhile, we designed an algorithmic framework based on 1D-CNN for feature extraction of multi-frequency vibration. We use the R-wave of ECG to truncate the vibration signals of multi-frequency bands to form many single-cycle waveforms, which are input into 1D-CNN to learn the feature point locations through training and finally output the feature point coordinates. Our study recognizes two feature points of PCG and eight feature points of SCG. Excluding the two feature points of RE and RF, the average
, RME, and RSME reach 0.95, 2.18 ms, and 4.89 ms, respectively, and have high recognition robustness against morphological variability. The average response time of the new method to process a single feature point is 60.18
which achieves low latency. Finally, combining the multi-frequency vibration model, acquisition sensor, and signal feature extraction method form a cardiac multi-frequency vibration dynamic monitoring system scheme. This scheme is a more efficient option that can be applied to portable acquisition devices for daily dynamic cardiac monitoring.
4. Discussion
This study first proposes a multi-frequency vibration model of the heart and designs the corresponding sensor and processing algorithms. The vibration model, the sensor, and feature extraction algorithms are integrated to form a multi-frequency vibration monitoring system for the heart. The system expands the frequency range of cardiac monitoring to include the ultra-low frequency band, infrasound band, and audible band, which can extract the comprehensive mechanical vibration information of the heart and combine it with the ECG to map more comprehensive cardiac information.
In our previous study, an electro-mechanical-acoustic activity model of the heart was proposed, emphasizing the association between electrical activity, low-frequency vibration, and high-frequency acoustic activity. Among them, low-frequency vibration and high-frequency acoustic activity are mechanically coupled as the system command response and subsequent higher harmonics of the heart pump, respectively [
22]. In this study, we further enrich the connotation of mechanical coupling and elaborate a theoretical model of multi-frequency vibration of the heart in combination with Fourier series theory. It is a new attempt to decompose the quasi-periodic vibration of the heart into vibration modes of different frequency band ranges according to the cardiac events: the fundamental frequency mode represented by ULF-SCG, the mid-frequency vibration mode represented by SCG, and the high-harmonic mode represented by PCG.
A multi-frequency vibration monitoring system for the heart is built based on vibration modeling, including sensor design and feature extraction algorithms. The sensors designed in this study can detect multi-frequency vibration signals compared to existing accelerometers, gyroscopes, etc. A comparison of multi-band vibration sensors is shown in
Table 8. Compared to existing sensors [
23,
24], it is more miniaturized and lightweight while satisfying high sensitivity. Compared to our previous study [
22], the sensor in this study becomes enhanced and optimized as the separate sensor structure is more compatible with the wearable design and the static pressure sensor is removed to obtain a smaller size. The 1D-CNN-based feature extraction framework proposed in this study can achieve high-accuracy extraction of multiple vibration feature points, including eight feature points of SCG and two feature points of PCG. A comparison of the feature extraction methods for SCG is shown in
Table 9. The method can recognize more feature points compared with the existing Gaussian mixture model [
26], high-frequency envelope [
27], and wavelet transform [
28]. Compared to the methods of binary classification [
29], it can ignore the morphological variability of the signal. Compared to the curvature method [
22], it can track the feature points of continuous signals and satisfy the low latency property, which can be applied to real-time monitoring.
The cardiac signals measured by the multi-frequency monitoring system are subject to motion artifacts and other disturbances, so we ensured that the subject remained stationary during the acquisition and applied a band-pass filter in the signal processing. It eliminates motion artifacts at low frequencies and noise at high frequencies to some extent for the current acquisition. Current applications are limited to daily stationary continuous monitoring. If we can eliminate interference during movement, we can expand the application to most everyday states, including active states such as walking and climbing stairs. Multi-channel detection would be potentially an effective way to eliminate motion interference [
11], where comparing signals from different channels and utilizing signal processing methods can eliminate the effects of motion interference. The sensor designed in this study meets the miniaturization characteristics and can meet the requirements of wearable morphology but, at present, the wearable functions of our devices have not been realized. To meet the requirement of being wearable, we need to solve the problems of wireless transmission, acquisition circuit integration, and power supply and at the same time realize the low power consumption and miniaturization design. For the 1D-CNN model, the current sample size of 1255 cannot cover all morphologies of waveforms, which will lead to a decrease in the recognition accuracy when confronted with new morphologies of waveforms. We can solve this problem gradually by increasing the sample size and waveform morphology, such as extensive data collection for healthy and non-healthy populations. In addition, the problem of insufficient sample size can be solved by increasing data diversity through data augmentation. It is also worth noting that the latency time test of the 1D-CNN model was performed on the above GPU computing platform. The delay time of the test includes model prediction and performance evaluation, excluding the time of data loading, and finally obtains an average prediction time of 60.18 us. Compared with the study of the same 1D-CNN method for estimating stress, the prediction time is 115.5 us/sample [
41], which can prove the low latency property of our proposed method. Meanwhile, the latency of algorithms running on embedded platforms changes because of the difference in arithmetic power. FPGA platforms are a good choice for embedded platforms because they can provide parallel computing. The compact CNN model clipped in this study can be implemented on FPGA and further performance enhancement can be conducted by computational kernel optimization, broadband optimization, etc. [
42,
43]. Our study demonstrates that the 1D-CNN method can obtain high-accuracy recognition results while satisfying the low latency property. Therefore, the latency test at this stage is reasonable.