MSIMG: A Density-Aware Multi-Channel Image Representation Method for Mass Spectrometry
Abstract
1. Introduction
2. Materials and Methods
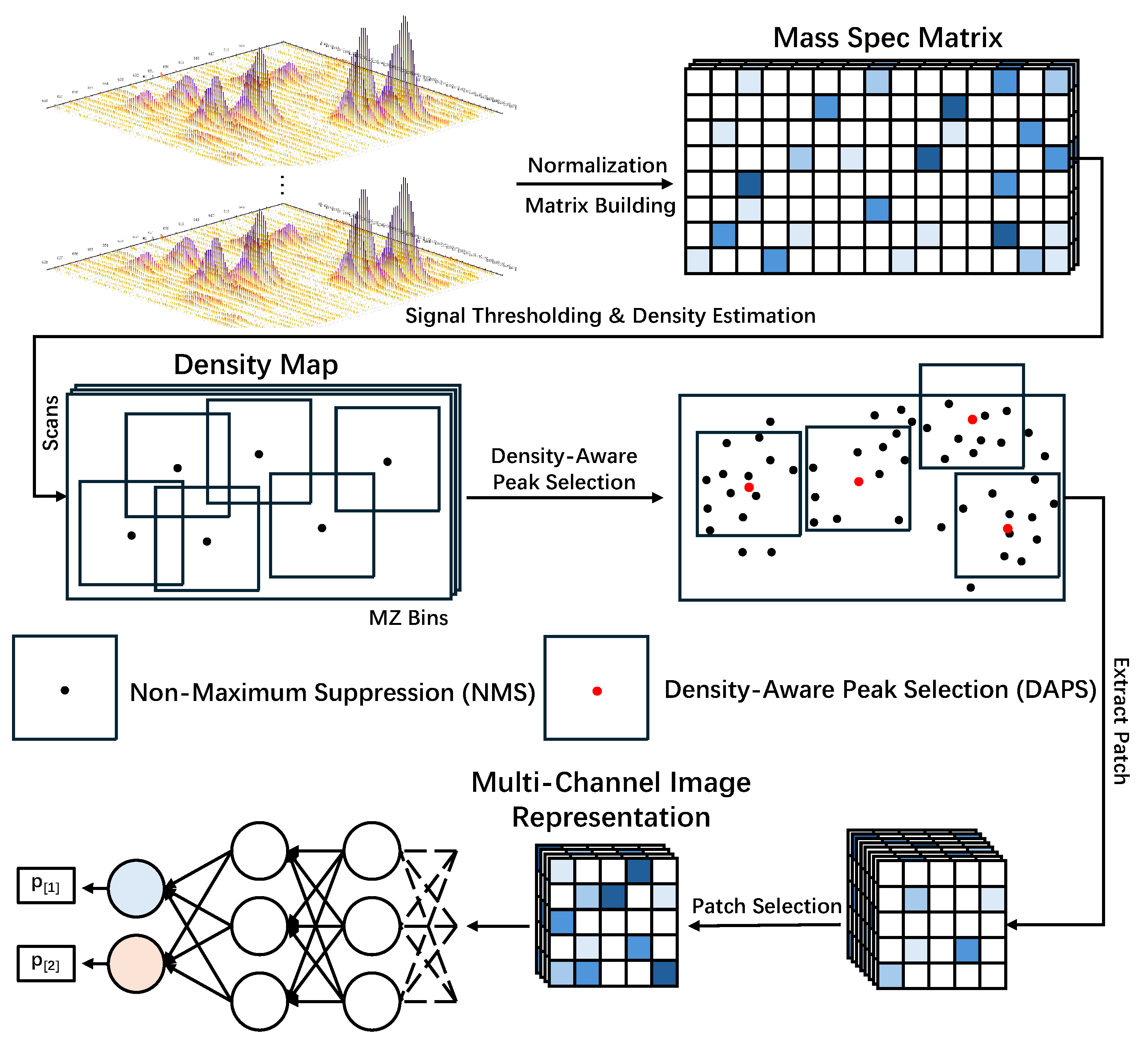
2.1. Mass Spec Matrix Construction
2.2. Density-Aware Peak Selection Strategy
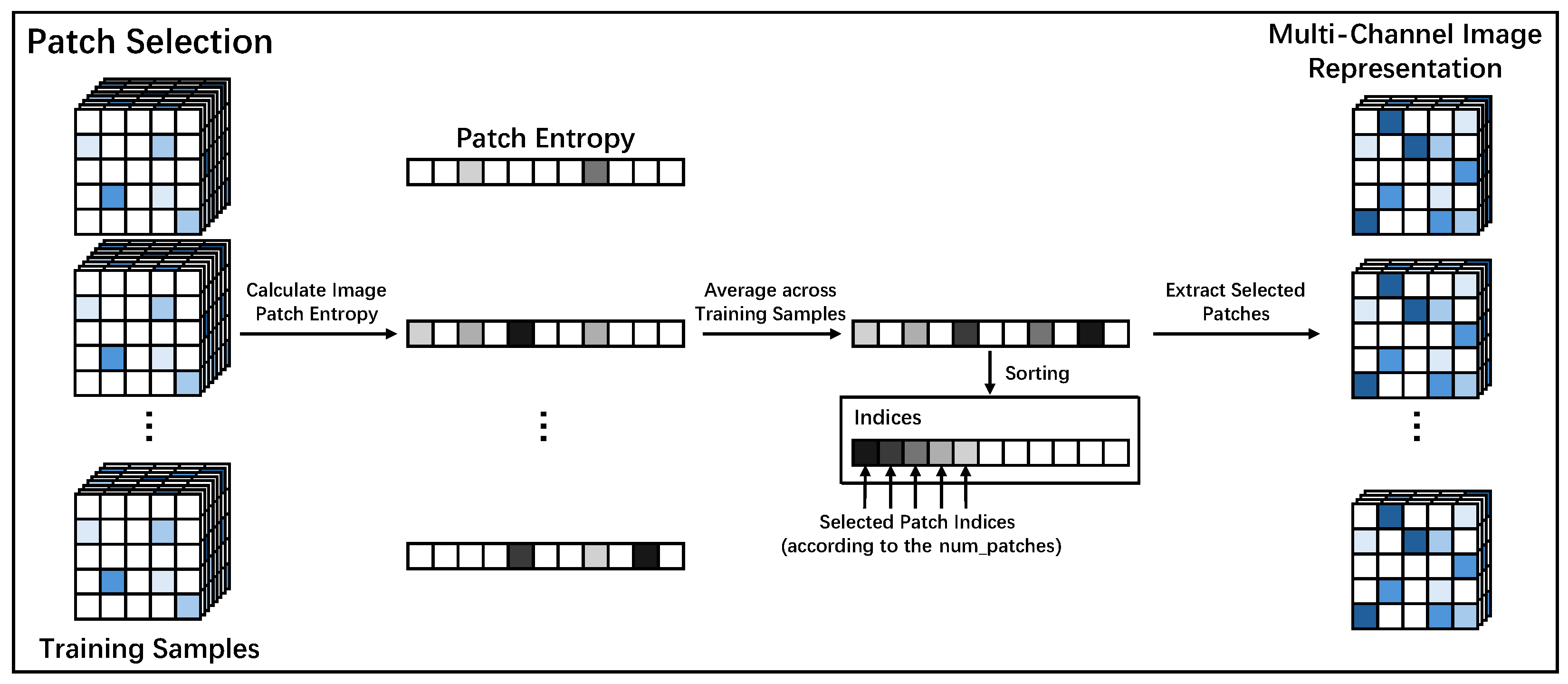
2.3. Construction of the Multi-Channel Image Representation
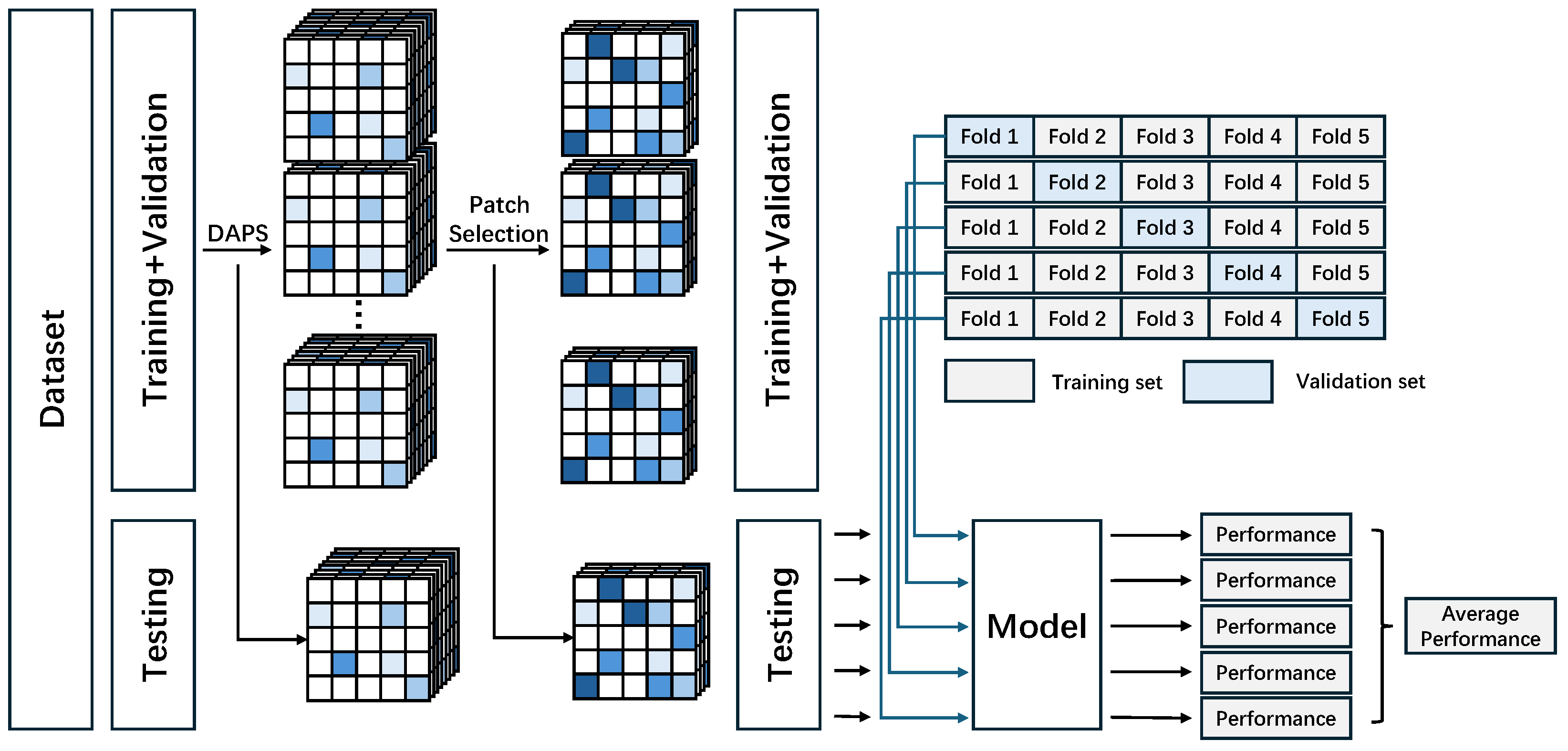
2.4. Model Training and Evaluation Strategy
2.5. Datasets
3. Results
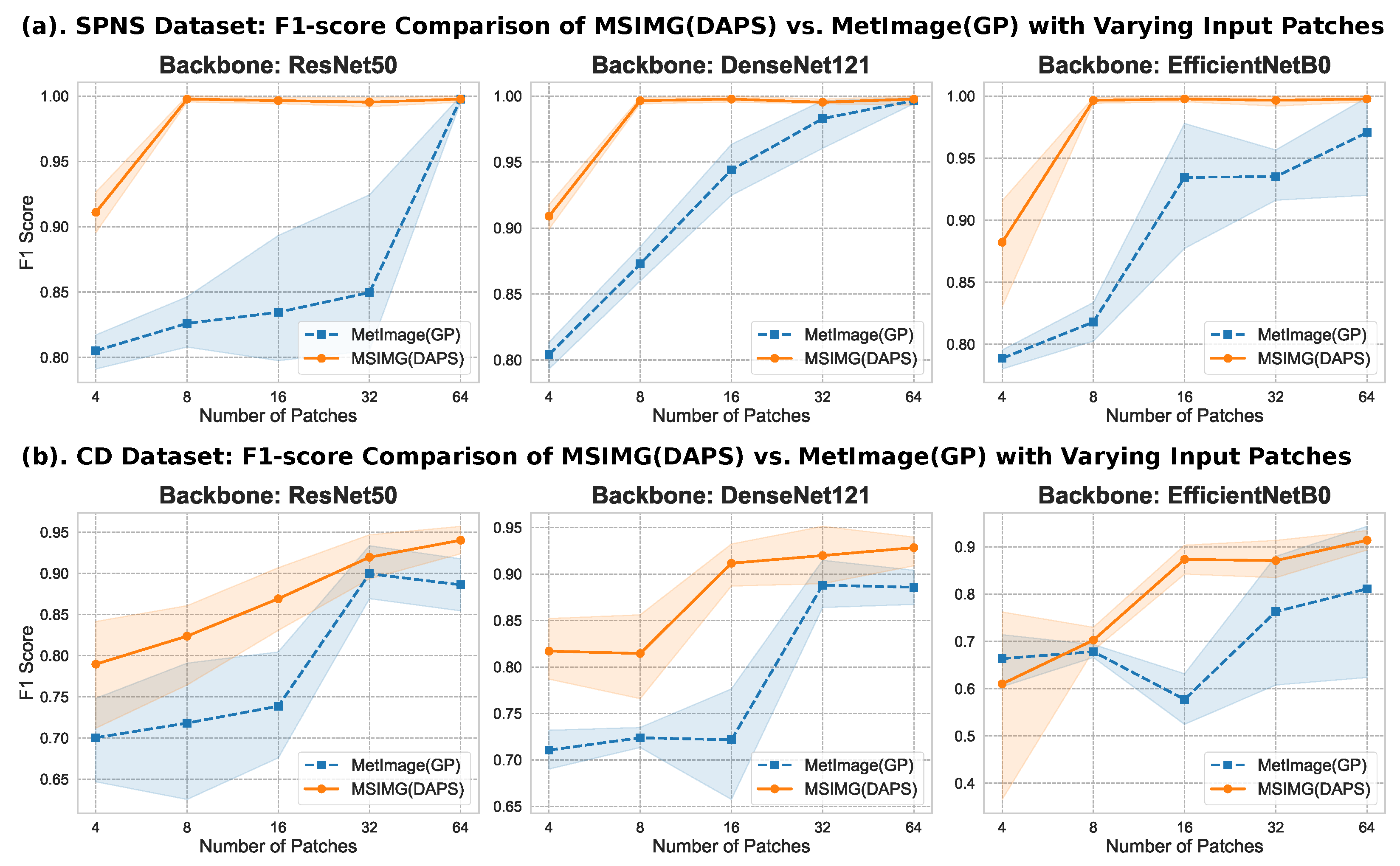
3.1. Parameter Sensitivity Analysis: Selection of Multi-Channel Image Dimensions and Number of Channels
3.2. Performance Comparison of Multi-Channel Image Representation and Traditional Methods
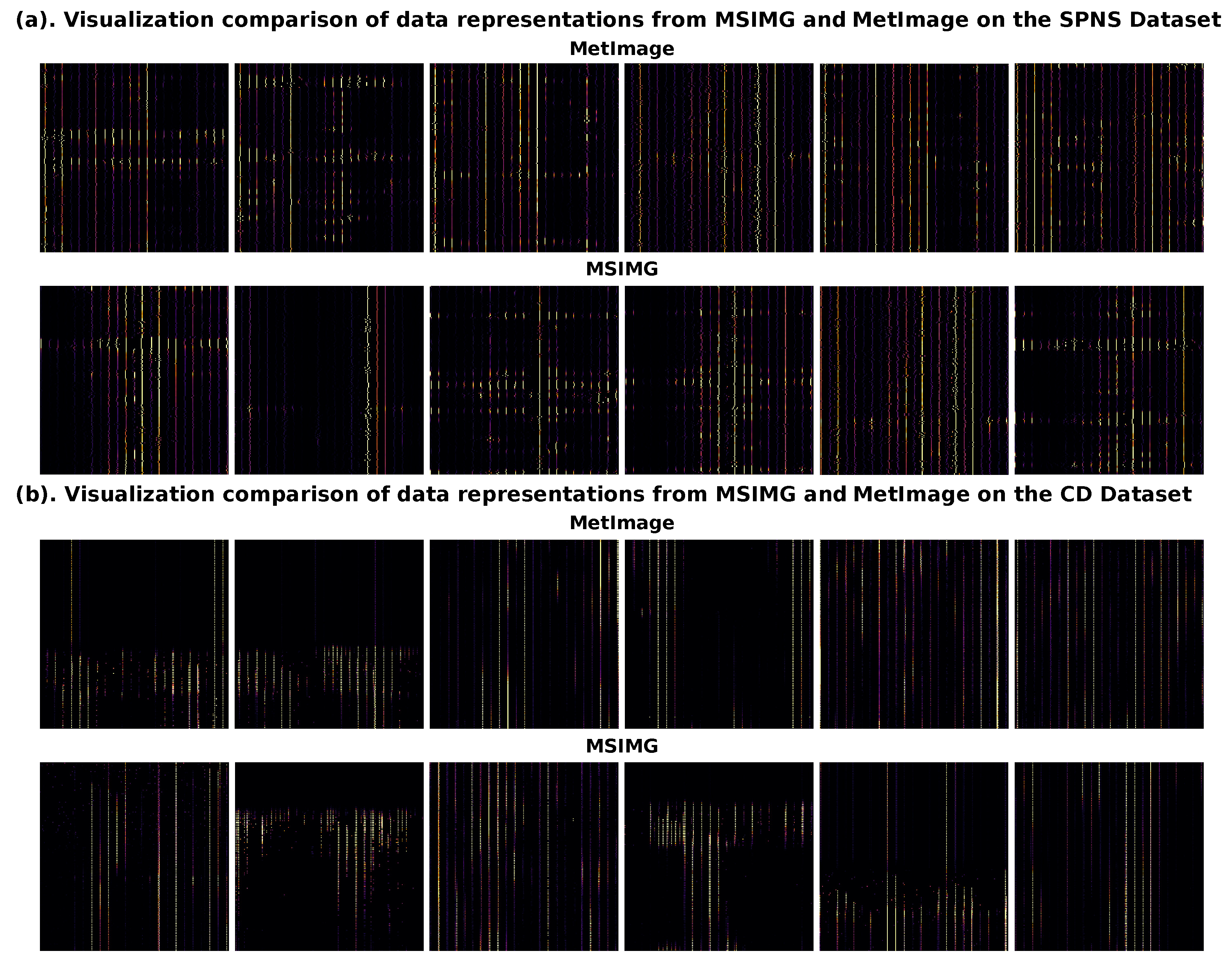
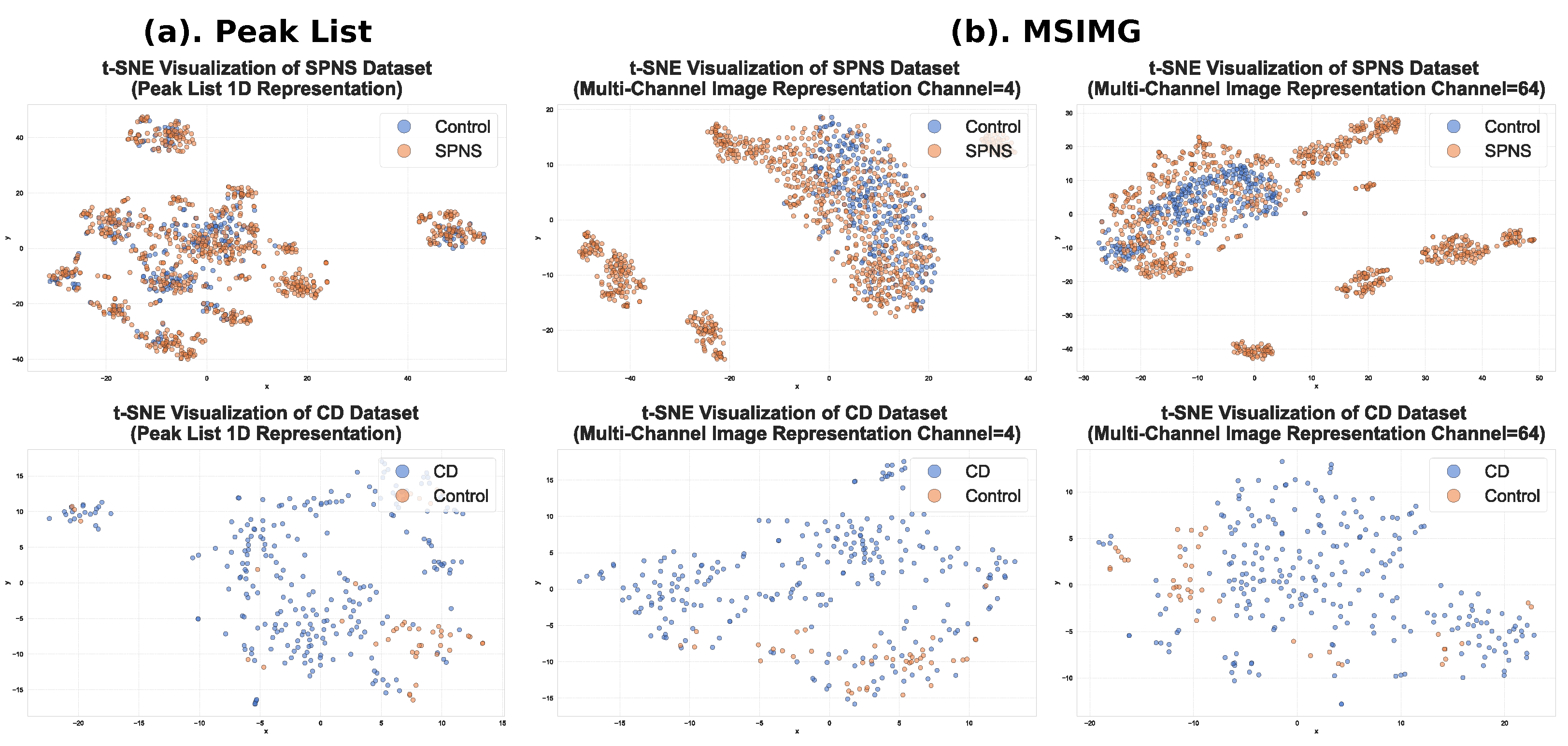
3.3. Visualization of the Effectiveness of Multi-Channel Image Representation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sumner, L.W.; Mendes, P.; Dixon, R.A. Plant metabolomics: Large-scale phytochemistry in the functional genomics era. Phytochemistry 2003, 62, 817–836. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Bedair, M.; Sumner, L.W. Current and emerging mass-spectrometry technologies for metabolomics. TrAC Trends Anal. Chem. 2008, 27, 238–250. [Google Scholar] [CrossRef]
- Lei, Z.; Huhman, D.V.; Sumner, L.W. Mass spectrometry strategies in metabolomics. J. Biol. Chem. 2011, 286, 25435–25442. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass spectrometry-based proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Aslanian, A.; Yates, J.R., III. Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 2008, 12, 483–490. [Google Scholar] [CrossRef]
- Zhou, X.X.; Zeng, W.F.; Chi, H.; Luo, C.; Liu, C.; Zhan, J.; He, S.M.; Zhang, Z. pDeep: Predicting MS/MS spectra of peptides with deep learning. Anal. Chem. 2017, 89, 12690–12697. [Google Scholar] [CrossRef]
- Petricoin, E.F.; Liotta, L.A. Mass spectrometry-based diagnostics: The upcoming revolution in disease detection. Clin. Chem. 2003, 49, 1227–1229. [Google Scholar] [CrossRef]
- Silva, C.J.; Onisko, B.C.; Dynin, I.; Erickson, M.L.; Requena, J.R.; Carter, J.M. Utility of mass spectrometry in the diagnosis of prion diseases. Anal. Chem. 2011, 83, 1609–1615. [Google Scholar] [CrossRef]
- Yoshida, M.; Hatano, N.; Nishiumi, S.; Irino, Y.; Izumi, Y.; Takenawa, T.; Azuma, T. Diagnosis of gastroenterological diseases by metabolome analysis using gas chromatography–mass spectrometry. J. Gastroenterol. 2012, 47, 9–20. [Google Scholar] [CrossRef] [PubMed]
- May, J.C.; McLean, J.A. Advanced multidimensional separations in mass spectrometry: Navigating the big data deluge. Annu. Rev. Anal. Chem. 2016, 9, 387–409. [Google Scholar] [CrossRef] [PubMed]
- Chaleckis, R.; Meister, I.; Zhang, P.; Wheelock, C.E. Challenges, progress and promises of metabolite annotation for LC–MS-based metabolomics. Curr. Opin. Biotechnol. 2019, 55, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. Imagenet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Cadow, J.; Manica, M.; Mathis, R.; Reddel, R.R.; Robinson, P.J.; Wild, P.J.; Hains, P.G.; Lucas, N.; Zhong, Q.; Guo, T.; et al. On the feasibility of deep learning applications using raw mass spectrometry data. Bioinformatics 2021, 37, i245–i253. [Google Scholar] [CrossRef]
- Huang, J.; Li, Y.; Meng, B.; Zhang, Y.; Wei, Y.; Dai, X.; An, D.; Zhao, Y.; Fang, X. ProteoNet: A CNN-based framework for analyzing proteomics MS-RGB images. iScience 2024, 27, 111362. [Google Scholar] [CrossRef]
- Shen, X.; Shao, W.; Wang, C.; Liang, L.; Chen, S.; Zhang, S.; Rusu, M.; Snyder, M.P. Deep learning-based pseudo-mass spectrometry imaging analysis for precision medicine. Brief. Bioinform. 2022, 23, bbac331. [Google Scholar] [CrossRef]
- Zhang, F.; Yu, S.; Wu, L.; Zang, Z.; Yi, X.; Zhu, J.; Lu, C.; Sun, P.; Sun, Y.; Selvarajan, S.; et al. Phenotype classification using proteome data in a data-independent acquisition tensor format. J. Am. Soc. Mass Spectrom. 2020, 31, 2296–2304. [Google Scholar] [CrossRef]
- Wang, H.; Yin, Y.; Zhu, Z.J. Encoding LC–MS-based untargeted metabolomics data into images toward AI-based clinical diagnosis. Anal. Chem. 2023, 95, 6533–6541. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, S.; He, B.; Sha, Q.; Shen, Y.; Yan, T.; Nian, R.; Lendasse, A. Gaussian derivative models and ensemble extreme learning machine for texture image classification. Neurocomputing 2018, 277, 53–64. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial Pyramid Pooling in Deep Convolutional Networks for Visual Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 37, 1904–1916. [Google Scholar] [CrossRef]
- Najibi, M.; Rastegari, M.; Davis, L.S. G-cnn: An iterative grid based object detector. In Proceedings of the IEEE conference on computer vision and pattern recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2369–2377. [Google Scholar]
- Lempitsky, V.; Zisserman, A. Learning to count objects in images. Adv. Neural Inf. Process. Syst. 2010, 23, 1324–1332. [Google Scholar]
- Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, PAMI-8, 679–698. [Google Scholar] [CrossRef]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Bentley, J.L. Multidimensional binary search trees used for associative searching. Commun. ACM 1975, 18, 509–517. [Google Scholar] [CrossRef]
- Zhou, W.; Lin, L.; Jiang, L.Y.; Wu, J.L.; Xu, W.C.; Zhou, Y.; Wang, M.J.; Cao, X.M.; Lin, H.Q.; Yang, J.; et al. Comprehensive plasma metabolomics and lipidomics of benign and malignant solitary pulmonary nodules. Metabolomics 2022, 18, 71. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning. PMLR, Long Beach, CA, USA, 9–15 June 2019; Chaudhuri, K., Salakhutdinov, R., Eds.; 2019; Volume 97, pp. 6105–6114. [Google Scholar]
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D.; et al. Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat. Methods 2021, 18, 747–756. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Cai, L.; Zhu, Y. The challenges of data quality and data quality assessment in the big data era. Data Sci. J. 2015, 14, 2. [Google Scholar] [CrossRef]
- Bengio, Y.; Courville, A.; Vincent, P. Representation learning: A review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef]

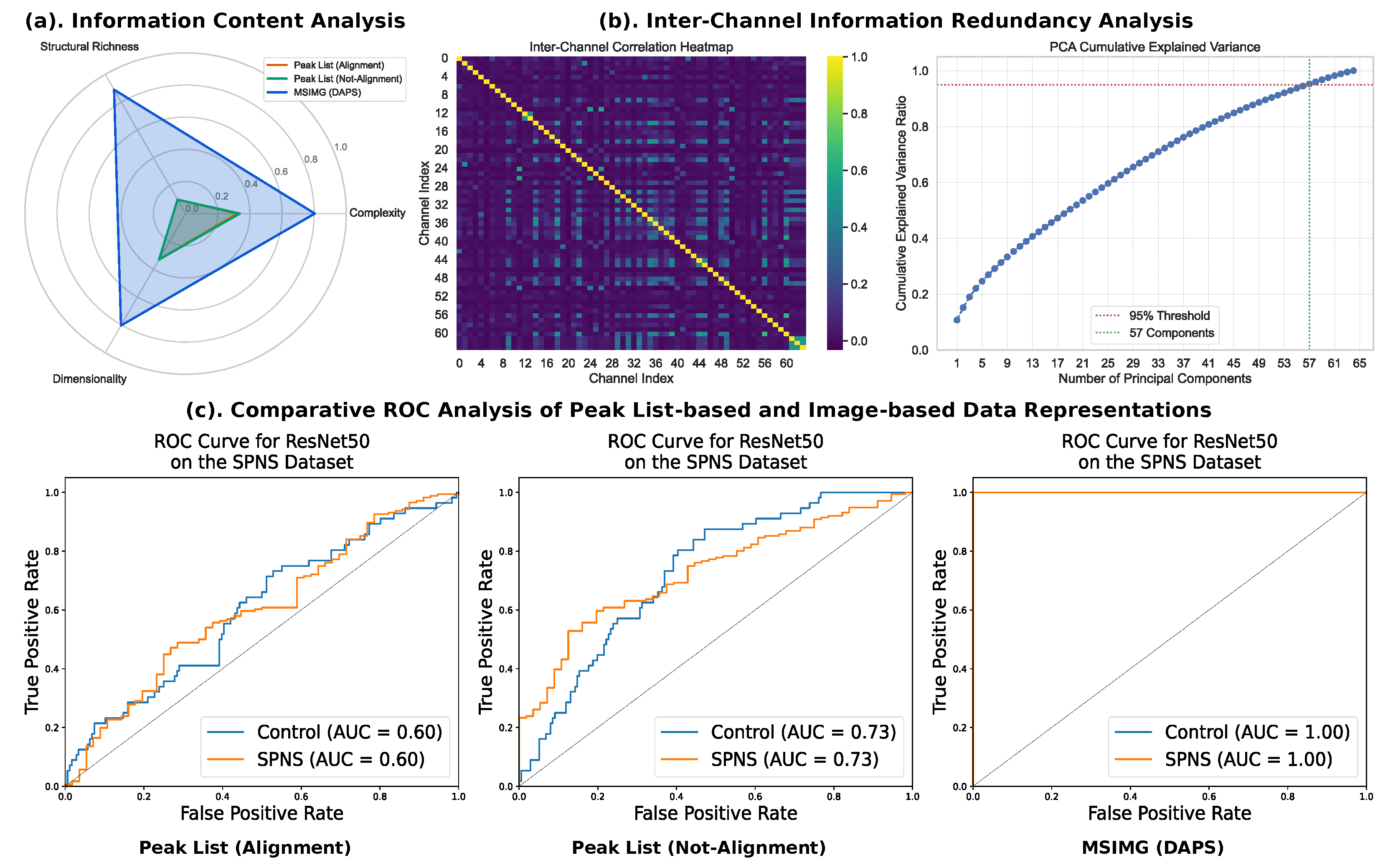
| Model | Peak List (Align) | Peak List (Not-Align) | MSIMG | |||
|---|---|---|---|---|---|---|
| Accuracy | F1 Score | Accuracy | F1 Score | Accuracy | F1 Score | |
| RF | 0.7819 ± 0.01 | 0.5563 ± 0.02 | 0.8129 ± 0.01 | 0.6671 ± 0.02 | - | - |
| SVM | 0.7604 ± 0.00 | 0.4387 ± 0.02 | 0.7595 ± 0.00 | 0.4351 ± 0.01 | - | - |
| LDA | 0.5414 ± 0.02 | 0.4905 ± 0.02 | 0.5586 ± 0.03 | 0.5126 ± 0.03 | - | - |
| ResNet50 | 0.7181 ± 0.05 | 0.4745 ± 0.05 | 0.6155 ± 0.03 | 0.5694 ± 0.02 | 0.9983 ± 0.00 | 0.9977 ± 0.00 |
| DenseNet121 | 0.6991 ± 0.08 | 0.4699 ± 0.05 | 0.6500 ± 0.02 | 0.5966 ± 0.02 | 0.9983 ± 0.00 | 0.9977 ± 0.01 |
| EfficientNetB0 | 0.7104 ± 0.06 | 0.4806 ± 0.05 | 0.6595 ± 0.04 | 0.5988 ± 0.03 | 0.9983 ± 0.00 | 0.9977 ± 0.00 |
| Model | Peak List (Align) | Peak List (Not-Align) | MSIMG | |||
|---|---|---|---|---|---|---|
| Accuracy | F1 Score | Accuracy | F1 Score | Accuracy | F1 Score | |
| RF | 0.8542 ± 0.02 | 0.5689 ± 0.05 | 0.9085 ± 0.02 | 0.7695 ± 0.05 | - | - |
| SVM | 0.8440 ± 0.01 | 0.5298 ± 0.04 | 0.8780 ± 0.01 | 0.6424 ± 0.06 | - | - |
| LDA | 0.7593 ± 0.04 | 0.6760 ± 0.04 | 0.8542 ± 0.04 | 0.7580 ± 0.05 | - | - |
| ResNet50 | 0.7424 ± 0.06 | 0.6238 ± 0.04 | 0.8542 ± 0.03 | 0.7677 ± 0.04 | 0.9661 ± 0.01 | 0.9402 ± 0.02 |
| DenseNet121 | 0.7864 ± 0.16 | 0.6817 ± 0.16 | 0.8237 ± 0.09 | 0.7556 ± 0.10 | 0.9593 ± 0.02 | 0.9284 ± 0.02 |
| EfficientNetB0 | 0.8339 ± 0.03 | 0.7035 ± 0.04 | 0.8237 ± 0.09 | 0.7355 ± 0.08 | 0.9492 ± 0.02 | 0.9138 ± 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Gao, B.; Wang, Y.; Guo, L.; Zhang, W.; Xiong, X. MSIMG: A Density-Aware Multi-Channel Image Representation Method for Mass Spectrometry. Sensors 2025, 25, 6363. https://doi.org/10.3390/s25206363
Zhang F, Gao B, Wang Y, Guo L, Zhang W, Xiong X. MSIMG: A Density-Aware Multi-Channel Image Representation Method for Mass Spectrometry. Sensors. 2025; 25(20):6363. https://doi.org/10.3390/s25206363
Chicago/Turabian StyleZhang, Fengyi, Boyong Gao, Yinchu Wang, Lin Guo, Wei Zhang, and Xingchuang Xiong. 2025. "MSIMG: A Density-Aware Multi-Channel Image Representation Method for Mass Spectrometry" Sensors 25, no. 20: 6363. https://doi.org/10.3390/s25206363
APA StyleZhang, F., Gao, B., Wang, Y., Guo, L., Zhang, W., & Xiong, X. (2025). MSIMG: A Density-Aware Multi-Channel Image Representation Method for Mass Spectrometry. Sensors, 25(20), 6363. https://doi.org/10.3390/s25206363






