Voltammetric Detection of Damage to DNA by Arsenic Compounds at a DNA Biosensor
Abstract
:1. Introduction
2. Experimental
2.1 Apparatus and Reagents
2.2 Procedure
3. Results and Discussion
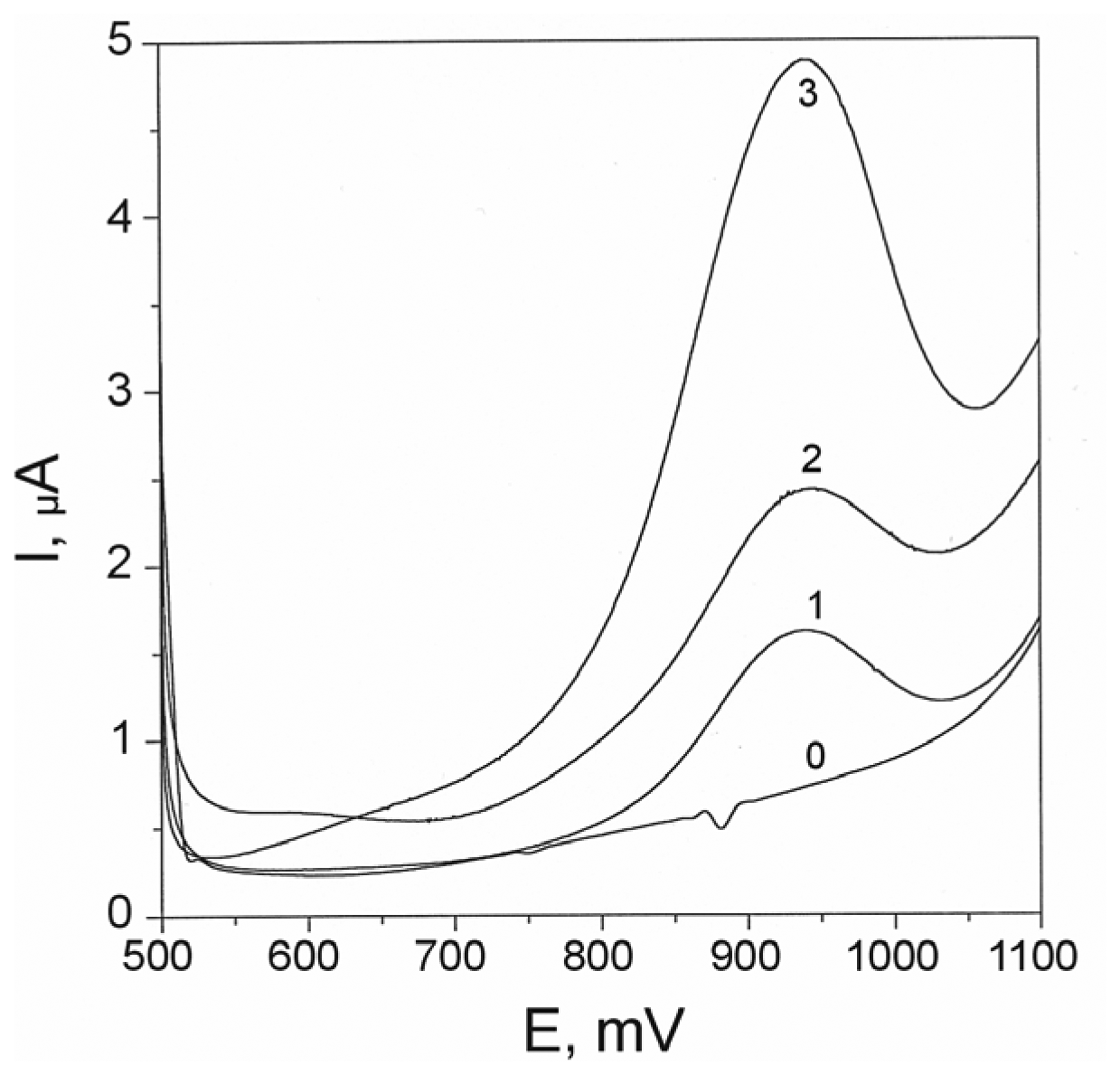
3.1 Optimization of detection conditions
3.2 Cleavage effect of inorganic arsenic
3.3 Cleavage effect of organic arsenic
Conclusions
Acknowledgments
References
- Palecek, E.; Fojta, M. Anal. Chem. 2001, 73, 74A–83A.
- Mascini, M.; Palchetti, I.; Marrazza, G. Fres. J. Anal. Chem. 2001, 369, 15–22.
- Wang, J. Anal. Chim. Acta 2002, 469, 63–71.
- Palecek, E.; Jelen, F. Crit. Rev. Anal. Chem. 2002, 32, 261–270.
- Ye, Y.; Ju, H. Sensors 2003, 3, 128–145.
- Wang, J. Anal. Chim. Acta 2003, 500, 247–257.
- Fojta, M. Collect. Czech. Chem. Commun. 2004, 69, 715–747.
- Labuda, J.; Fojta, M.; Jelen, F.; Paleček, E. Encyclopedia of Sensors; Grimes, C. A., Dickey, E., Eds.; American Scientidic Publishers: Stevenson Ranch, CA, in press.
- Abreu, F.C.; Goulart, M. O. F.; Oliveira Brett, A. M. Biosens. Bioelectron. 2002, 17, 913–919.
- Oliveira Brett, A. M.; da Silva, L. A.; Fujii, H.; Mataka, S.; Thiemann, T. J. Electroanal. Chem. 2003, 549, 91–99.
- Erdem, A.; Ozsoz, M. Electroanalysis 2002, 14, 965–974.
- Labuda, J.; Bučková, M.; Heilerová,, L'.; Šilhár, S.; Štepánek, I. Anal. Bioanal. Chem. 2003, 376, 168–173.
- Bode, A.M.; Dong, Z. Crit. Rev. Oncol../Hematol. 2002, 42, 5–24.
- Harris, G.K.; Shi, X. Mutation Res. 2003, 533, 183–200.
- Okayasu, R.; Takahashi, S.; Sato, H.; Kubota, Y.; Scolavino, S.; Bedford, J.S. DNA Repair 2003, 2, 309–314.
- Hartwig, A.; Blessing, H.; Schwerdtle, T.; Walter, I. Toxicol. 2003, 193, 161–169.
- Rossman, T.G. Mutation Res. 2003, 533, 37–65.
- Ng, J.C.; Wang, J.; Shraim, A. Chemosphere 2003, 52, 1353–1359.
- Gomez-Caminer, A.; Howe, P.; Hughes, M.; Kenyon, E.; Lewis, D. R.; Moore, M.; Ng, J.; Aitio, A.; Becking, G. Arsenic and Arsenic Compouns. In Environ.Health Criteria 224; WHO: Geneva, 2001. [Google Scholar]
- Hughes, M. F. Toxicol. Lett. 2002, 133, 1–16.
- Ahmad, S.; Kitchin, K. T.; Cullen, W. R. Arch. Biochem. Biophys. 2000, 382, 195–202.
- Labuda, J.; Bučková, M.; Vaníčková, M.; Mattusch, J.; Wennrich, R. Electroanalysis 1999, 11, 101–107.
- Fojta, M.; Staňková, V.; Paleček, E.; Koscielniak, P.; Mitáš, J. Talanta 1998, 46, 155–161.
- Ozsoz, M.; Erdem, A.; Kara, P.; Kerman, K.; Ozkan, D. Electroanalysis 2003, 15, 613–619.
- Pang, D. W.; Abruña, H. D. Anal. Chem. 1998, 70, 3162–3169.
- Dollimore, L. S.; Gillard, R. D. J. Chem. Soc. 1973, 78, 933–940.
- Labuda, J.; Bučková, M.; Heilerová,, L'.; Čaniová- Žiaková, A.; Brandšteterová, E.; Mattusch, J; Wennrich, R. Sensors 2002, 2, 1–10.



| Cleavage mixture | DNA marker signal (I/I0) | |||
|---|---|---|---|---|
| Composition | Concentration, mol/L | No As | As(III) | As(V) |
| Fe(II) H2O2 | 2×10-4 1.5×10-2 | 0.90±0.05 | - | - |
| As Fe(II) H2O2 | 6×10-3 2×10-4 1.5×10-2 | - | 0.83±0.03 | 0.90±0.04 |
| As Fe(II) H2O2, 37 °C | 6×10-3 2×10-4 1.5×10-2 | - | 0.94±0.03 | - |
| As Fe(III) H2O2 | 6×10-3 2×10-4 1.5×10-2 | - | 0.99±0.02 | - |
| Cleavage mixture | DNA marker signal (I/I0) | |||
|---|---|---|---|---|
| Composition | Concentration, mol/L | No As | As(III) | As(V) |
| Cu(II) H2O2 | 2×10-4 1.5×10-2 | 0.92±0.11 | - | - |
| As Cu(II) H2O2 | 6×10-3 2×10-4 1.5×10-2 | - | 0.79±0.08 | 0.95±0.02 |
| Cu(II) H2O2, 37 °C | 2×10-7 1.5×10-2 | 0.95±0.07 | - | - |
| As Cu(II) H2O2, 37 °C | 6×10-3 2×10-7 1.5×10-2 | - | 1.21±0.04 | - |
| Cleavage mixture | DNA marker signal (I/I0) | |||
|---|---|---|---|---|
| Composition | Concentration, mol/L | DMA | PhA | APhA |
| As | 1×10-3 | 0.90±0.01 | 1.00±0.05 | 0.76±0.04 |
| As Fe(II) H2O2 | 1×10-3 2×10-4 1.5×10-2 | 0.58±0.04 | 0.66±0.11 | 0.61±0.05 |
| As Cu(II) H2O2 | 1×10-3 2×10-4 1.5×10-2 | 0.67±0.08 | 0.73±0.03 | 0.83±0.03 |
© 2005 by MDPI ( http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Labuda, J.; K. Bubnicova, K.; Kovalova, L.; Vanickova, M.; Mattusch, J.; Wennrich, R. Voltammetric Detection of Damage to DNA by Arsenic Compounds at a DNA Biosensor. Sensors 2005, 5, 411-423. https://doi.org/10.3390/s5060411
Labuda J, K. Bubnicova K, Kovalova L, Vanickova M, Mattusch J, Wennrich R. Voltammetric Detection of Damage to DNA by Arsenic Compounds at a DNA Biosensor. Sensors. 2005; 5(6):411-423. https://doi.org/10.3390/s5060411
Chicago/Turabian StyleLabuda, J., K. K. Bubnicova, L. Kovalova, M. Vanickova, J. Mattusch, and R. Wennrich. 2005. "Voltammetric Detection of Damage to DNA by Arsenic Compounds at a DNA Biosensor" Sensors 5, no. 6: 411-423. https://doi.org/10.3390/s5060411
APA StyleLabuda, J., K. Bubnicova, K., Kovalova, L., Vanickova, M., Mattusch, J., & Wennrich, R. (2005). Voltammetric Detection of Damage to DNA by Arsenic Compounds at a DNA Biosensor. Sensors, 5(6), 411-423. https://doi.org/10.3390/s5060411




