Abstract
Primary malignancies of the lung, skin (melanoma), and breast have higher propensity for metastatic spread to the brain. Advances in molecular tumour profiling have aided the development of targeted therapies, stereotactic radiotherapy, and immunotherapy, which have led to some improvement in patient outcomes; however, the overall prognosis remains poor. Continued research to identify new prognostic and predictive biomarkers is necessary to further impact patient outcomes, as this will enable better risk stratification at the point of primary cancer diagnosis, earlier detection of metastatic deposits (for example, through surveillance), and more effective systemic treatments. Brain metastases exhibit considerable inter- and intratumoural heterogeneity—apart from distinct histology, treatment history and other clinical factors, the metastatic brain tumour microenvironment is incredibly variable both in terms of subclonal diversity and cellular composition. This review discusses emerging biomarkers; specifically, the biological context and potential clinical utility of tumour tissue biomarkers, circulating tumour cells, extracellular vesicles, and circulating tumour DNA.
1. Introduction
Brain cancers can be primary, arising within different areas of the brain, or metastatic, arising from different organs of the body and spreading to the brain, also known as brain metastasis (BrM). This review focuses on discussing BrM-related biomarkers. These metastatic tumours are most frequent in lung cancers, followed by breast, melanoma, colon, kidney, and ovarian cancers, and 15% of cases with unknown primary origin [1]. Over the past 40 years, the quality of life and outcomes of patients with BrM have improved, owing to progress in neuroimaging, neurosurgery, and oncology. Magnetic resonance imaging is a standard procedure for diagnosis. BrMs usually appear as contrast-enhancing lesions, most frequently in the cerebral hemispheres (80% of cases), cerebellum (15%), and brainstem (<5%) [2]. Under a newly revised graded prognostic assessment [3], BrM patient survival rates have improved; however, they are still variable. For lung cancer, median overall survival is 15 months, 14–16 months for breast, 7–10 months for melanoma, 5–8 months for GI cancers, and 10–12 months for renal cell carcinoma [4]. This variability reflects the heterogenous patient population and highlights the complexities of clinical management.
2. Current Clinical Management Strategies for Brain Metastases
The management of BrM patients is usually coordinated in a multidisciplinary team setting, with treatment planning based on patient age, Karnofsky performance status (a general well-being scale), the primary origin of the cancer as some primaries have a higher risk of BrM, and the extent of extracranial disease control [5,6]. Corticosteroids and antiepileptic agents are routinely used for neurological symptom management. In terms of local treatment, neurosurgical tumour resection is considered depending on the location and the number of metastases within the brain (oligometastatic; limited metastatic burden or highly metastatic), with contemporary surgical interventions such as cortical mapping [7] and laser interstitial thermal therapy [8] used to minimize perioperative morbidity. Irradiating the whole brain remains a critical option when surgery is not appropriate, but stereotactic radiosurgery (SRS) is preferred for patients with up to or more than four BrM [9] and good performance status because it targets radiation very precisely to minimize treatment-induced neurocognitive side effects.
Various guidelines are available for treatment of brain metastasis from solid tumours, including the European Association of Neuro-Oncology (EANO)–European Society for Medical Oncology (ESMO) [10] and the National Comprehensive Cancer network (NCCN) [11]. Guidelines from the NCCN define BrM patients to be treated with surgery, stereotactic radiosurgery (SRS), whole-brain radiation therapy (WBRT), and systemic therapy [11]. However, new recommendations for patients presenting with extensive brain metastasis, usually with a known history of cancer, undergo either resection or biopsy to confirm CNS involvement, and then they may be subjected to WBRT, SRS, or systemic therapy and then follow-up with brain MRI every 2–3 months for the next 1–2 years. NCCN encourages that patients with limited metastatic lesions should undergo a prior consultation phase with a multidisciplinary team to optimise the best treatment plan. For instance, patients with disseminated systemic disease with poor response may undergo WBRT or SRS or perhaps consider palliative care. Conversely, patients with newly diagnosed or stable systemic disease may undergo SRS or WBRT. Nonetheless, then these patients will be followed up by brain MRI every 2–3 months for 1–2 years and every 4–6 months indefinitely. Furthermore, depending on either recurrence locally or in distant sites, they might be evaluated for further surgery, SRS, WBRT, and systemic therapy [11]. The systemic therapy regimen usually depends on the primary tumour, for instance, BrM patients with breast primaries are treated with breast cancer regimen on the basis of the breast cancer subtype, lung BrM patients are given lung cancer regimens, and so on. Overall, systemic therapy such as chemotherapeutics such as cisplatin, paclitaxel, and temozolomide have shown mixed results in clinical trials [12,13]; however, they are still used in practice if they can help stabilize extracranial metastases and they may have an impact in the brain in some patients. Any prescription of molecular-targeted therapy is based on primary histology, treatment history, and data on actionable alterations where available. For example, trastuzumab is used for HER2-amplified breast cancer BrM, tragisso (Osimertinib) for EGFR-mutant lung cancer BrM [14,15], and xalkori (Crizotinib), alecensa (Alectinib), alunbrig (Brigatinib), and zykadia (Ceritinib) against ALK-rearranged BrM [4,15,16,17,18,19,20,21]. However, molecular-targeted agents are generally also given with the goal of stabilising, rather than eliminating, metastatic deposits, as complete intracranial responses are rare. Factors that limit the efficacy of the existing treatments include intratumoural heterogeneity, insufficient vascular permeability due to the blood–brain barrier (BBB), elevated tumour interstitial fluid pressure, and other aspects of the tumour microenvironment (TME) [22,23,24,25].
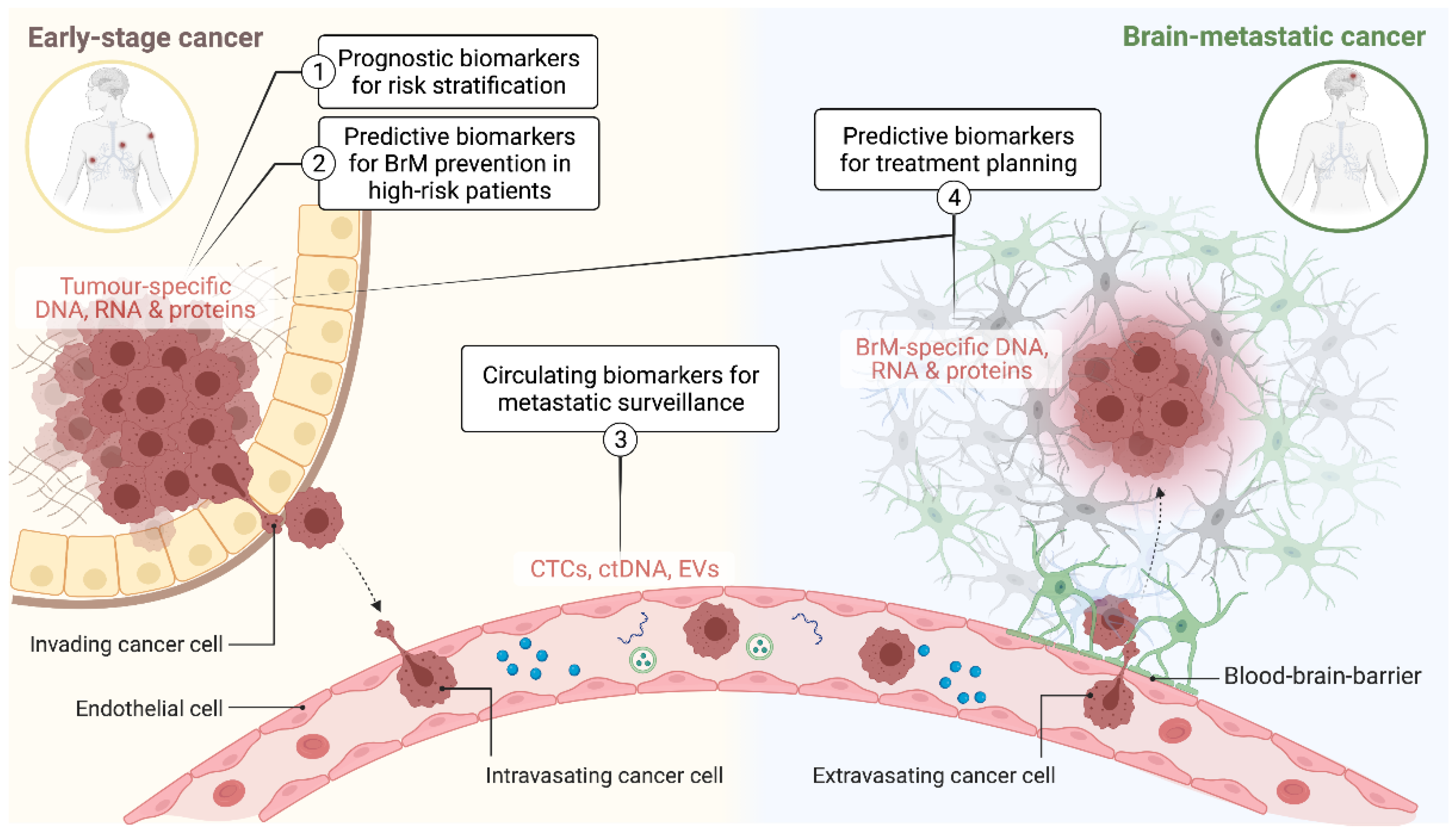
For new experimental agents, increasing the number of phase II and III trials that specifically test intracranial efficacy as a secondary endpoint will be important if we are to increase the number of options available in the brain metastatic setting [26,27]. Developing companion biomarkers in parallel to molecular target discovery and drug development is an essential component of this (Figure 1). This includes diagnostic tools for more accurate risk stratification and earlier intervention for low-stage disease (prognostic biomarkers), earlier detection, and monitoring of metastatic disease load (surveillance biomarkers) and treatment planning (predictive biomarkers). This article comprehensively revises evidence supporting the potential clinical utility and/or biological context of BrM biomarkers, highlighting the most recent advances in both pre-clinical and clinical settings.
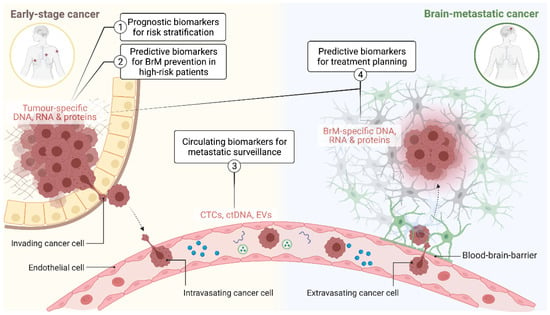
Figure 1.
Schematic illustrating the application of predictive and prognostic biomarkers at different stages of brain metastasis pathogenesis. CTCs: circulating tumour cells; ctDNA: circulating tumour DNA; EVs: extracellular vesicles; BrM: brain metastasis.
3. Biomarkers for Prognostication and Differential Diagnosis of BrM
Measuring changes in the expression of individual proteins or multi-gene signatures has dominated the
landscape of prognostic biomarker research. These studies often share a common
goal of being able to predict the risk of BrM at the point of primary cancer
diagnosis. For example, Duchnowska identified a 13 gene BrM prediction
signature for HER2+ breast cancer [28], and this was further refined to
a 3 gene classifier that included RAD51 (RAD51 homolog), HDGF (hepatoma-derived
growth factor), and TPR (translocated promoter region) genes.
Interestingly, multivariate analysis revealed that this 3 gene signature was
highly predictive of early BrM in the discovery cohort, but it was not
confirmed in the validation cohort. These results indicate that significant
differences between patient cohorts may impact the genes expressed that drive
the metastatic pattern. There were differences in the linic-pathological
characteristics, treatment regimens, and number of patients, which may have
resulted in the non-confirmation of the signature. Nonetheless, it identified
few crucial genes which might lead to BrM development. Kamer et al. discovered
a similarly indicated signature for lung cancer, which additionally predicted
metastatic spread with and without brain involvement [29]. The
genes involved metastatic spread to the brain from the lungs were mainly
oxidative phosphorylation pathway genes, indicating that perhaps in BrM, a more
efficient way of utilising glucose which is highly available in this
microenvironment is necessary to meet the demands of the rapidly growing
metastasis. Prognostic protein biomarkers have also been explored, offering the
key practical advantage of rapid integration into current clinical diagnostic
practice as most laboratories already have cost-efficient processes for
immunohistochemistry (IHC). For example, expression of NDRG1 (N-myc
downregulated gene 1) is higher in BrM-initiating breast tumour cells [30], and expression
of PLEKHA5 (Pleckstrin homology domain-containing A5) in melanoma is associated
with brain-specific metastasis [31]. The detection of markers such
as PLEKHA5 and NDRG1 in primary tumour biopsies could provide an early
indication of aggressive phenotype, providing they can be fully validated.
Practical limitations on prognostic biomarker translation are expertly reviewed
elsewhere [32,33].
To be implemented in clinical diagnostic practice, biomarker discoveries require validation in independent sample cohorts, reaching acceptable levels of sensitivity, specificity, and feasibility in a single sample assay context, as well as favourable cost–benefit analysis. Few biomedical research discoveries proceed to this stage, but those that do tend to focus on patient subpopulations where clinical decision making is not optimally served by existing diagnostic frameworks. One such group is BrM patients whose primary tumour type cannot be unequivocally identified by existing differential diagnosis approaches. The frequency of cancers of unknown primary (CUP) may be as high as 16% of all BrM in some institutions [34]. Patients with no history of prior malignancy potentially benefit the most from accurate diagnostic information at this stage, as in theory, their disease is still sensitive to first-line agents.
Genomic mutation profiling is often used for CUP diagnosis, especially in patients with widespread metastatic disease, providing a BrM tissue sample is available. For example, epidermal growth factor receptor (EGFR) mutations and ALK rearrangements are usually indicative of a lung cancer origin (most CUP cases), while BRAF and NRAS mutations are associated with melanoma. EGFR mutations are also prognostic after BrM diagnosis [35,36]. Multigene signatures may have a role, providing there is infrastructure for these assays. Zheng et al. developed a 90 gene signature that distinguished between primary and metastatic brain tumours with 99% accuracy, and for BrM, correctly predicted primary tumour histology 89% of the time [37].
Cerebrospinal fluid (CSF) is gaining credibility for brain tumour diagnostic applications [38], with some studies reporting that biomarkers might be more abundant in CSF than peripheral fluids [39,40,41,42]. Next-generation sequencing of ctDNA from CSF of 53 patients revealed that >50% of samples harboured somatic alterations specific to brain metastatic disease [43]. Other studies have also demonstrated the diagnostic utility of CSF ctDNA analysis, with mutation profiles generally consistent with the primary tumour type (e.g., NRAS and BRAF mutations from melanoma patients and EGFR mutations for lung BrM patients [42]). These studies highlight the potential of CSF as a biomarker identification medium, but CSF sampling is considered highly invasive. It may be a suitable source of biomarkers only if other circulating biomarkers are uninformative. It may also be possible that CSF provides BrM-specific diagnostic information, but this is yet to be demonstrated in a clinical setting.
4. Surveillance Biomarkers
The majority of BrMs are diagnosed at an advanced stage when patients already have neurological symptoms. If there are also other comorbidities from extracranial disease, this poses major challenges for disease management, and the primary goal of treatment in this setting is to stabilise, rather than cure, disease. Current diagnostic tools are unable to account for tumour heterogeneity or track progression without multiple, lengthy imaging appointments. More efficient alternatives are needed to enable regular monitoring and expedite early screening. A non-invasive mode of disease monitoring that is gaining momentum is the regular screening of whole blood or blood products for circulating biomarkers, including circulating tumour cells (CTCs), cell-free DNA (cfDNA), and extracellular vesicles [44,45,46]. These are further discussed below, and Table 1 lists the specific emerging biomarkers that may be relevant for early identification of BrM, or for monitoring disease load after treatment has been initiated.

Table 1.
Various biomarkers and their sources with applications as predictive and prognostic biomarkers of brain metastasis. The advantages and disadvantages have also been listed [35,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74].
4.1. Circulating Tumour Cells (CTCs)
CTCs are tumour cells shed from a solid mass into the circulation (blood and lymph). They are thought to be the opportunistic ’seeds’ of metastatic progression capable of forming micro-metastatic reservoirs [75,76,77]. Evidence indicates their systemic load correlates roughly with the extent of spread, making CTCs a relevant biomarker for monitoring systemic disease load [78]. In 2018, Hanssen explored the predictive value of CTCs in lung cancer patients with oligometastatic brain disease, reporting a significantly worse prognosis for patients with a higher CTC load [79]. The main challenge translating this knowledge into a clinical practice has been the quantification of CTCs, due to their low overall count in the blood. Various in vitro and in vivo strategies are being tested to overcome this, but it remains a persistent barrier to clinical implementation.
Currently, there is just one commercially available, FDA-approved platform for CTC counting—CellSearch®, manufactured by Veridex. It uses an antibody against epithelial cell adhesion molecule (EpCAM), which is present on carcinoma cells but not on normal blood cells [80]. The commercial platform does not detect EpCAM-negative CTCs [81], but Huebner recently developed a filtration-based system with better sensitivity [82]. It involves filtering CTC subpopulations and then sorting them by flow cytometry, an approach that seemed to have greater prognostic value for metastatic breast cancer patients with overall low CTC load. This filtration system is based on an automated nucleic acid preparation system (VERSANT® kPCR), which can also simultaneously purify DNA, RNA, or proteins from CTCs for further analysis.
Single-cell CTC profiling has highlighted metastasis-specific alterations that could be targeted to maximise sensitivity [83,84]. For example, in a study of breast BrM patients, Boral et al. identified exclusive elevated Ki67 expression as well as other cell death and immune evasion pathways in BrM-CTCs compared to non-BrM CTCs [50]. The differential expression of these pathways in BrM-specific CTCs highlight potential applications in detection of early stage BrM.
4.2. Circulating Cell-Free DNA
Circulating, cell-free DNA (cfDNA) released from tumour cells may be a powerful biomarker of BrM. Its abundance increases as the integrity of the BBB is progressively compromised in expanding tumours. In contrast to CTCs where sensitivity is often an issue, a major consideration for cfDNA-based surveillance is specificity—distinguishing cfDNA from normal and tumour cells. Circulating tumour DNA (ctDNA; specifically shed from tumour cells) constitutes around 1% of total cfDNA and can be distinguished on the basis of size and genetic profile. ctDNA fragments are shorter than cfDNA from normal cells and harbour somatic mutations [85,86], making ctDNA an ideal diagnostic biomarker. Furthermore, it can be readily detected in plasma or cerebrospinal fluid using droplet digital PCR (ddPCR)—a precise, sensitive, and robust technique that is also relatively user-friendly for the clinical diagnostic setting.
In early stage melanoma, serum lactate dehydrogenase (LDH) is routinely assayed because early intervention based on elevated LDH improves patient outcomes. Using ddPCR to detect BRAF or NRAS mutations in ctDNA of metastatic melanoma patients, Chang et al. found that mutated ctDNA was elevated in 83% of patients with BrM, whereas LDH was elevated in only 50% of cases. Hence, at least in this study, ctDNA had significantly higher sensitivity than LDH for monitoring disease progression [87]. With respect to clinical translation, a randomized trial with paired CT scans and ctDNA plasma sampling in bevacizumab-treated patients with non-resectable metastatic melanoma is ongoing (NCT02872259). Because of the reduced life expectancy, CNS involvement is an exclusion criterion that limits the study to extracranial disease monitoring. Nonetheless, melanoma BrM patients are still likely to benefit from a ddPCR-based cfDNA test being implemented, even if it is on the basis of extracranial monitoring data.
4.3. Extracellular Vesicles
Extracellular vesicles (EVs) are small membrane-bound organelles that are released from cells. They transport different cargoes through the circulation, including proteins, nucleic acids, lipids, miRNAs, and metabolites. Exosomes are small EVs involved in cell–cell communication. As a substrate for disease monitoring, exosomes are becoming more mainstream rather than a niche research field. According to a market report from Grand View Research, exosome research and their clinical use will be worth USD 2.28 billion by 2030, representing an annual growth rate of 18.8% [88]. Several preclinical studies reported a crucial role for exosomes in brain colonisation and the BrM TME [61,72,89,90,91,92]. EVs are isolated via density gradient ultracentrifugation or size-exclusion chromatography. Both are specialised and labour-intensive, being unsuitable for a clinical diagnostic laboratory. Newer technologies that circumvent the current practical limitations will be critical for implementation in the clinic, as will standardized protocols for isolation and analysis.
BrM-derived exosomes that contribute to BBB dysfunction through miRNA-181c may be a substrate for surveillance, as serum miR-181c is elevated in metastatic breast cancer patients with BrM [92]. Similar studies have reported EV or exosome-mediated tumour progression, but their potential for surveillance has not been investigated. For example, EV-derived miRNA-122, involved in metabolic reprogramming [91], exosomal miR-19a released by activated astrocytes in the BrM TME [61], and exosomal CEMIP (cell migration inducing hyaluronidase 1) protein whose uptake by brain endothelial and microglial cells induces pro-inflammatory cytokines and vascular remodelling [72].
5. Predictive Biomarkers for Treatment Planning
Predicting treatment responses in central nervous system tumours is highly valuable and the beneficial outcomes of this has been widely discussed elsewhere [93,94,95,96]. Overall precision cancer medicine is an evolving but accepted new norm in many parts of the developed world [97,98]. The paradigm is supported by basket trials that established the benefits of treating cancer patients on the basis of their specific mutation profile rather than histology alone [99]. BrM patients can benefit from precision care in centres where this is routine, although neurosurgical tissue is not always available. It is possible that predictive information could be garnered from a liquid biopsy, but the feasibility and accuracy of this route have not yet been demonstrated. A primary tumour biopsy may be the next best option at present, with the caveats that BrM outgrowth potentially not being dependent on targetable alterations identified in the primary, and that BrM-specific or clonal alterations may be missed.
Brastianos et al. performed whole-exome sequencing on 86 BrM and matching primary tumour samples, predominantly from breast, lung, and kidney cancer patients. The divergent genetic profiles of sample pairs illuminated the dynamics of clonal evolution during progression, but also highlighted possibilities to personalise the prescription of molecular-targeted therapy, with BrM samples frequently harbouring alterations that confer sensitivity to PI3K/AKT/mTOR, CDK, and HER2/EGFR inhibitors [100]. Our own genomic analysis of Australian cases concurred, with actionable mutations identified in 86% of BrM analysed [101]. A more recent study by Tyran and colleagues confirmed the benefits of genomic testing on BrM samples rather than primary tumours, where available [102].
Another potential target in BrM is homologous recombination deficiency (HRD). Most often caused by pathogenic mutations in BRCA1 and BRCA2, HRD compromises the repair of double-stranded DNA breaks, exacerbating overall genomic instability and increasing the dependence on poly-ADP-ribose polymerase (PARP)-dependent repair. Exploiting a synthetic lethality between HRD and BRCA1/2, PARP inhibitors such as olaparib are now used to treat familial ovarian and breast tumours with this genotype [103,104]. In metastatic breast cancer, HRD was found to be higher in BrM than the primary tumour [105]. Therefore, the use of PARP inhibitors in the BrM setting could be an important ancillary application worth investigating. Other studies revealed elevated HRD and mismatch repair deficiency signatures in BrMs compared to matching primary breast and colorectal cancers [102,106].
Human epidermal growth family receptors (HER) have been extensively studied in BrMs by our group and others [101,107,108,109]. HER3 and HER4 activation are elevated in BrM, but not EGFR, suggesting this is a microenvironment-driven feature since neuregulins are abundant in the brain but EGF is not [101,107,108]. Similarly, RNA sequencing of longitudinally collected BrM from breast cancer revealed elevated RET and HER2 signalling. Their inhibition reduced proliferation in patient-derived tissue cultures and significantly slowed the growth of matching patient-derived xenograft (PDX) models [110]. Hence, TME-dependent changes could be exploited by dual targeting of these proteins along with PI3K inhibitors [109].
The advent of immunotherapy targeting immunomodulatory proteins such as programmed cell death protein 1 receptor (PD-1) and its ligands has shown promise in many cancers [111,112]. With respect to BrMs, a phase II clinical trial on melanoma BrM patients [113] and phase I on NSCLC BrM patients [114] showed the combined use of ipilimumab and nivolumab resulted in an improved clinical response compared to individual monotherapies. Despite this breakthrough, many patients do not benefit from immunotherapy. Over the last few years, focus has shifted to deciphering predictive biomarkers for the efficacy of immune checkpoint inhibitors (ICIs). For example, with continued advancement of multiplex immunohistochemical technology, as well as high-throughput sequencing, an array of multifactorial predictive markers can be developed [94]. Tumour mutation burden (TMB) test is an adopted guideline by the NCCN for patients with NSCLC receiving immunotherapy. For NSCLC, tumour mutation burden is site-specific, and lung BrMs have the highest TMB. Metastatic sites in lung adenocarcinomas generally had higher TMB with increased PD-L1 positivity [115]. This raises the possibility of investigating ICI treatment in a site-specific manner on the basis of high TMB for NSCLC. However, TMB is not the best predictor of ICI response for many cancers [116]. It should be noted that PD-1/PD-L1 expression and TMB cut-offs may vary across studies, thereby presenting as a challenge to standardize the cut-off criteria for future applications. Furthermore, the feasibility and reproducibility of standardized predictive biomarkers would have to be established to leverage this towards precision immune-oncology for BrMs.
6. Conclusions and Perspectives
The clinical management of BrM patients is a complex challenge that would benefit greatly from more precise diagnostic information at multiple stages of disease progression. Investment should be primarily focused on the development of superior biomarkers for early stage cancer that help prevent BrM in more patients and reduce overall rates of distant relapse, particularly companion diagnostics (predictive markers) that accurately predict the response to individual therapies. There is also great potential for non-invasive monitoring of early cancer patients deemed to be at high risk of BrM (e.g., through regular blood sampling), as this provides an opportunity to intercept newly metastatic patients as early as possible, improving the likelihood of a complete response to second-line therapy and improving patient quality of life. Those with established metastatic disease who are undergoing treatment also stand to benefit from real-time surveillance via blood sampling because it has the potential to be more sensitive and cost-effective than re-staging by PET/MR imaging. There is a very strong rationale for additional research and development on surveillance biomarkers, as well as technology that helps to overcome the practical barriers currently blocking the realisation of this approach in clinical practice.
Author Contributions
Conceptualization, P.K.-d.C.; writing—original draft preparation, P.K.-d.C., V.J.; writing—review and editing, P.K.-d.C., V.J., J.M.S., S.R.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant from the Australian National Health and Medical Research Council (APP1017028).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The scope for this review was broad, and we apologise to authors whose work we could not cite due to space constraints.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Achrol, A.S.; Rennert, R.C.; Anders, C.; Soffietti, R.; Ahluwalia, M.S.; Nayak, L.; Peters, S.; Arvold, N.D.; Harsh, G.R.; Steeg, P.S.; et al. Brain metastases. Nat. Rev. Dis. Primers 2019, 5, 5. [Google Scholar] [CrossRef]
- Lin, B.; Huang, D.; Yang, X.; Zhang, Y.; Gang, F.; Du, X.B. Distribution of brain metastases: Low-risk metastasis areas may be avoided when treating with whole-brain radiotherapy. Cancer Imaging 2020, 20, 29. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. Neuro-Oncology 2020, 22, iv1–iv96. [Google Scholar] [CrossRef] [PubMed]
- Sperduto, P.W.; Mesko, S.; Li, J.; Cagney, D.; Aizer, A.; Lin, N.U.; Nesbit, E.; Kruser, T.J.; Chan, J.; Braunstein, S.; et al. Survival in Patients With Brain Metastases: Summary Report on the Updated Diagnosis-Specific Graded Prognostic Assessment and Definition of the Eligibility Quotient. J. Clin. Oncol. 2020, 38, 3773–3784. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, L.; Scott, C.; Rotman, M.; Asbell, S.; Phillips, T.; Wasserman, T.; McKenna, W.G.; Byhardt, R. Recursive partitioning analysis (RPA) of prognostic factors in three Radiation Therapy Oncology Group (RTOG) brain metastases trials. Int. J. Radiat. Oncol. Biol. Phys. 1997, 37, 745–751. [Google Scholar] [CrossRef]
- Gaspar, L.E.; Scott, C.; Murray, K.; Curran, W. Validation of the RTOG recursive partitioning analysis (RPA) classification for brain metastases. Int. J. Radiat. Oncol. Biol. Phys. 2000, 47, 1001–1006. [Google Scholar] [CrossRef]
- Sanmillan, J.L.; Fernandez-Coello, A.; Fernandez-Conejero, I.; Plans, G.; Gabarros, A. Functional approach using intraoperative brain mapping and neurophysiological monitoring for the surgical treatment of brain metastases in the central region. J. Neurosurg. 2017, 126, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, A.; McNichols, R.J.; Stafford, R.J.; Guichard, J.P.; Reizine, D.; Delaloge, S.; Vicaut, E.; Payen, D.; Gowda, A.; George, B. Laser thermal therapy: Real-time MRI-guided and computer-controlled procedures for metastatic brain tumors. Lasers Surg. Med. 2011, 43, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Serizawa, T.; Shuto, T.; Akabane, A.; Higuchi, Y.; Kawagishi, J.; Yamanaka, K.; Sato, Y.; Jokura, H.; Yomo, S.; et al. Stereotactic radiosurgery for patients with multiple brain metastases (JLGK0901): A multi-institutional prospective observational study. Lancet Oncol. 2014, 15, 387–395. [Google Scholar] [CrossRef]
- Le Rhun, E.; Guckenberger, M.; Smits, M.; Dummer, R.; Bachelot, T.; Sahm, F.; Galldiks, N.; de Azambuja, E.; Berghoff, A.S.; Metellus, P.; et al. EANO-ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up of patients with brain metastasis from solid tumours. Ann. Oncol. 2021, 32, 1332–1347. [Google Scholar] [CrossRef]
- Nabors, L.B.; Portnow, J.; Ahluwalia, M.; Baehring, J.; Brem, H.; Brem, S.; Butowski, N.; Campian, J.L.; Clark, S.W.; Fabiano, A.J.; et al. Central Nervous System Cancers, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2020, 18, 1537–1570. [Google Scholar] [CrossRef]
- Cortes, J.; Rodriguez, J.; Aramendia, J.M.; Salgado, E.; Gurpide, A.; Garcia-Foncillas, J.; Aristu, J.J.; Claver, A.; Bosch, A.; Lopez-Picazo, J.M.; et al. Front-line paclitaxel/cisplatin-based chemotherapy in brain metastases from non-small-cell lung cancer. Oncology 2003, 64, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Antonadou, D.; Paraskevaidis, M.; Sarris, G.; Coliarakis, N.; Economou, I.; Karageorgis, P.; Throuvalas, N. Phase II randomized trial of temozolomide and concurrent radiotherapy in patients with brain metastases. J. Clin. Oncol. 2002, 20, 3644–3650. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Ahn, M.J.; Garassino, M.C.; Han, J.Y.; Katakami, N.; Kim, H.R.; Hodge, R.; Kaur, P.; Brown, A.P.; Ghiorghiu, D.; et al. CNS Efficacy of Osimertinib in Patients With T790M-Positive Advanced Non-Small-Cell Lung Cancer: Data From a Randomized Phase III Trial (AURA3). J. Clin. Oncol. 2018, 36, 2702–2709. [Google Scholar] [CrossRef] [PubMed]
- Reungwetwattana, T.; Nakagawa, K.; Cho, B.C.; Cobo, M.; Cho, E.K.; Bertolini, A.; Bohnet, S.; Zhou, C.; Lee, K.H.; Nogami, N.; et al. CNS Response to Osimertinib Versus Standard Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Patients With Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2018, JCO2018783118. [Google Scholar] [CrossRef] [PubMed]
- Sperduto, P.W.; Jiang, W.; Brown, P.D.; Braunstein, S.; Sneed, P.; Wattson, D.A.; Shih, H.A.; Bangdiwala, A.; Shanley, R.; Lockney, N.A.; et al. Estimating Survival in Melanoma Patients With Brain Metastases: An Update of the Graded Prognostic Assessment for Melanoma Using Molecular Markers (Melanoma-molGPA). Int. J. Radiat. Oncol. Biol. Phys. 2017, 99, 812–816. [Google Scholar] [CrossRef]
- Sperduto, P.W.; Yang, T.J.; Beal, K.; Pan, H.; Brown, P.D.; Bangdiwala, A.; Shanley, R.; Yeh, N.; Gaspar, L.E.; Braunstein, S.; et al. Estimating Survival in Patients With Lung Cancer and Brain Metastases: An Update of the Graded Prognostic Assessment for Lung Cancer Using Molecular Markers (Lung-molGPA). JAMA Oncol. 2017, 3, 827–831. [Google Scholar] [CrossRef]
- Sperduto, P.W.; Kased, N.; Roberge, D.; Xu, Z.; Shanley, R.; Luo, X.; Sneed, P.K.; Chao, S.T.; Weil, R.J.; Suh, J.; et al. Effect of tumor subtype on survival and the graded prognostic assessment for patients with breast cancer and brain metastases. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, 2111–2117. [Google Scholar] [CrossRef]
- Sperduto, P.W.; Yang, T.J.; Beal, K.; Pan, H.; Brown, P.D.; Bangdiwala, A.; Shanley, R.; Yeh, N.; Gaspar, L.E.; Braunstein, S.; et al. The Effect of Gene Alterations and Tyrosine Kinase Inhibition on Survival and Cause of Death in Patients With Adenocarcinoma of the Lung and Brain Metastases. Int. J. Radiat. Oncol. Biol. Phys. 2016, 96, 406–413. [Google Scholar] [CrossRef]
- Kotecha, R.; Miller, J.A.; Venur, V.A.; Mohammadi, A.M.; Chao, S.T.; Suh, J.H.; Barnett, G.H.; Murphy, E.S.; Funchain, P.; Yu, J.S.; et al. Melanoma brain metastasis: The impact of stereotactic radiosurgery, BRAF mutational status, and targeted and/or immune-based therapies on treatment outcome. J. Neurosurg. 2018, 129, 50–59. [Google Scholar] [CrossRef]
- Miller, J.A.; Kotecha, R.; Ahluwalia, M.S.; Mohammadi, A.M.; Suh, J.H.; Barnett, G.H.; Murphy, E.S.; Vogelbaum, M.A.; Angelov, L.; Chao, S.T. The impact of tumor biology on survival and response to radiation therapy among patients with non-small cell lung cancer brain metastases. Pract. Radiat. Oncol. 2017, 7, e263–e273. [Google Scholar] [CrossRef]
- Arvanitis, C.D.; Ferraro, G.B.; Jain, R.K. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat. Rev. Cancer 2020, 20, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.; Puttick, S.; Houston, Z.H.; Thurecht, K.J.; Kalita-de Croft, P.; Mahler, S.; Rose, S.E.; Jeffree, R.L.; Mazzieri, R.; Dolcetti, R.; et al. Innovative Therapeutic Strategies for Effective Treatment of Brain Metastases. Int. J. Mol. Sci. 2019, 20, 1280. [Google Scholar] [CrossRef] [PubMed]
- Croft, P.K.-d.; Chittoory, H.; Nguyen, T.H.; Saunus, J.M.; Kim, W.G.; Reed, A.E.M.; Lim, M.; De Luca, X.M.; Ferguson, K.; Niland, C.; et al. Characterization of Immune Cell Subsets of Tumor Infiltrating Lymphocytes in Brain Metastases. Biology 2021, 10, 425. [Google Scholar] [CrossRef]
- Kalita-de Croft, P.; Straube, J.; Lim, M.; Al-Ejeh, F.; Lakhani, S.R.; Saunus, J.M. Proteomic Analysis of the Breast Cancer Brain Metastasis Microenvironment. Int. J. Mol. Sci. 2019, 20, 2524. [Google Scholar] [CrossRef] [PubMed]
- Alexander, B.M.; Brown, P.D.; Ahluwalia, M.S.; Aoyama, H.; Baumert, B.G.; Chang, S.M.; Gaspar, L.E.; Kalkanis, S.N.; Macdonald, D.R.; Mehta, M.P.; et al. Clinical trial design for local therapies for brain metastases: A guideline by the Response Assessment in Neuro-Oncology Brain Metastases working group. Lancet Oncol. 2018, 19, e33–e42. [Google Scholar] [CrossRef]
- Fabi, A.; Vidiri, A. Defining the endpoints: How to measure the efficacy of drugs that are active against central nervous system metastases. Transl. Lung Cancer Res. 2016, 5, 637–646. [Google Scholar] [CrossRef]
- Duchnowska, R.; Jassem, J.; Goswami, C.P.; Gokmen-Polar, Y.; Li, L.; Thorat, M.A.; Flores, N.; Hua, E.; Woditschka, S.; Palmieri, D.; et al. 13-gene signature to predict rapid development of brain metastases in patients with HER2-positive advanced breast cancer. J. Clin. Oncol. 2012, 30, 505. [Google Scholar] [CrossRef]
- Kamer, I.; Steuerman, Y.; Daniel-Meshulam, I.; Perry, G.; Izraeli, S.; Perelman, M.; Golan, N.; Simansky, D.; Barshack, I.; Ben Nun, A.; et al. Predicting brain metastasis in early stage non-small cell lung cancer patients by gene expression profiling. Transl. Lung Cancer Res. 2020, 9, 682–692. [Google Scholar] [CrossRef]
- Berghoff, A.S.; Liao, Y.; Karreman, M.A.; Ilhan-Mutlu, A.; Gunkel, K.; Sprick, M.R.; Eisen, C.; Kessler, T.; Osswald, M.; Wünsche, S. Identification and characterization of cancer cells that initiate metastases to the brain and other organs. Mol. Cancer Res. 2021, 19, 688–701. [Google Scholar] [CrossRef]
- Jilaveanu, L.B.; Parisi, F.; Barr, M.L.; Zito, C.R.; Cruz-Munoz, W.; Kerbel, R.S.; Rimm, D.L.; Bosenberg, M.W.; Halaban, R.; Kluger, Y. PLEKHA5 as a biomarker and potential mediator of melanoma brain metastasis. Clin. Cancer Res. 2015, 21, 2138–2147. [Google Scholar] [CrossRef]
- Simon, R. Lost in translation: Problems and pitfalls in translating laboratory observations to clinical utility. Eur. J. Cancer 2008, 44, 2707–2713. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miquel-Cases, A.; Schouten, P.C.; Steuten, L.M.; Retèl, V.P.; Linn, S.C.; van Harten, W.H. (Very) Early technology assessment and translation of predictive biomarkers in breast cancer. Cancer Treat. Rev. 2017, 52, 117–127. [Google Scholar] [CrossRef]
- Pekmezci, M.; Perry, A. Neuropathology of brain metastases. Surg. Neurol. Int. 2013, 4, S245–S255. [Google Scholar] [CrossRef]
- Li, W.-Y.; Zhao, T.-T.; Xu, H.-M.; Wang, Z.-N.; Xu, Y.-Y.; Han, Y.; Song, Y.-X.; Wu, J.-H.; Xu, H.; Yin, S.-C. The role of EGFR mutation as a prognostic factor in survival after diagnosis of brain metastasis in non-small cell lung cancer: A systematic review and meta-analysis. BMC Cancer 2019, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.A.; Milevskiy, M.J.; Lee, W.C.; Curry, M.C.; Smart, C.E.; Saunus, J.M.; Reid, L.; Da Silva, L.; Marcial, D.L.; Dray, E. The calcium pump plasma membrane Ca(2+)-ATPase 2 (PMCA2) regulates breast cancer cell proliferation and sensitivity to doxorubicin. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Zheng, Y.; Ding, Y.; Wang, Q.; Sun, Y.; Teng, X.; Gao, Q.; Zhong, W.; Lou, X.; Xiao, C.; Chen, C.; et al. 90-gene signature assay for tissue origin diagnosis of brain metastases. J. Transl. Med. 2019, 17, 331. [Google Scholar] [CrossRef] [PubMed]
- Bertero, L.; Siravegna, G.; Rudà, R.; Soffietti, R.; Bardelli, A.; Cassoni, P. Review: Peering through a keyhole: Liquid biopsy in primary and metastatic central nervous system tumours. Neuropathol. Appl. Neurobiol. 2019, 45, 655–670. [Google Scholar] [CrossRef]
- Martinez-Ricarte, F.; Mayor, R.; Martinez-Saez, E.; Rubio-Perez, C.; Pineda, E.; Cordero, E.; Cicuendez, M.; Poca, M.A.; Lopez-Bigas, N.; Ramon, Y.C.S.; et al. Molecular Diagnosis of Diffuse Gliomas through Sequencing of Cell-Free Circulating Tumor DNA from Cerebrospinal Fluid. Clin. Cancer Res. 2018, 24, 2812–2819. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Mayor, R.; Ng, C.K.Y.; Weigelt, B.; Martinez-Ricarte, F.; Torrejon, D.; Oliveira, M.; Arias, A.; Raventos, C.; Tang, J.; et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun. 2015, 6, 8839. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Pan, C.; Diplas, B.H.; Chen, X.; Wu, Y.; Xiao, X.; Jiang, L.; Geng, Y.; Xu, C.; Sun, Y.; Zhang, P.; et al. Molecular profiling of tumors of the brainstem by sequencing of CSF-derived circulating tumor DNA. Acta Neuropathol. 2019, 137, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Pentsova, E.I.; Shah, R.H.; Tang, J.; Boire, A.; You, D.; Briggs, S.; Omuro, A.; Lin, X.; Fleisher, M.; Grommes, C.; et al. Evaluating Cancer of the Central Nervous System Through Next-Generation Sequencing of Cerebrospinal Fluid. J. Clin. Oncol. 2016, 34, 2404–2415. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef]
- Loke, S.Y.; Lee, A.S.G. The future of blood-based biomarkers for the early detection of breast cancer. Eur. J. Cancer 2018, 92, 54–68. [Google Scholar] [CrossRef]
- Hanash, S.M.; Baik, C.S.; Kallioniemi, O. Emerging molecular biomarkers—Blood-based strategies to detect and monitor cancer. Nat. Rev. Clin. Oncol. 2011, 8, 142. [Google Scholar] [CrossRef]
- Klotz, R.; Thomas, A.; Teng, T.; Han, S.M.; Iriondo, O.; Li, L.; Restrepo-Vassalli, S.; Wang, A.; Izadian, N.; MacKay, M.; et al. Circulating Tumor Cells Exhibit Metastatic Tropism and Reveal Brain Metastasis Drivers. Cancer Discov. 2020, 10, 86. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Aceto, N.; Bersani, F.; Madden, M.W.; Donaldson, M.C.; Desai, R.; Zhu, H.; Comaills, V.; Zheng, Z.; et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014, 345, 216. [Google Scholar] [CrossRef]
- Pierga, J.Y.; Bidard, F.C.; Cropet, C.; Tresca, P.; Dalenc, F.; Romieu, G.; Campone, M.; Mahier Ait-Oukhatar, C.; Le Rhun, E.; Goncalves, A.; et al. Circulating tumor cells and brain metastasis outcome in patients with HER2-positive breast cancer: The LANDSCAPE trial. Ann. Oncol. 2013, 24, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Boral, D.; Vishnoi, M.; Liu, H.N.; Yin, W.; Sprouse, M.L.; Scamardo, A.; Hong, D.S.; Tan, T.Z.; Thiery, J.P.; Chang, J.C. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Zhao, W.; Lu, C.; Liu, D.; Li, P.; Ye, X.; Zhao, Y.; Zhang, J.; Yang, D. Next-Generation Sequencing Analysis of ctDNA for the Detection of Glioma and Metastatic Brain Tumors in Adults. Front. Neurol. 2020, 11, 544. [Google Scholar] [CrossRef]
- Seoane, J.; De Mattos-Arruda, L.; Le Rhun, E.; Bardelli, A.; Weller, M. Cerebrospinal fluid cell-free tumour DNA as a liquid biopsy for primary brain tumours and central nervous system metastases. Ann. Oncol. 2019, 30, 211–218. [Google Scholar] [CrossRef]
- Seremet, T.; Jansen, Y.; Planken, S.; Njimi, H.; Delaunoy, M.; El Housni, H.; Awada, G.; Schwarze, J.K.; Keyaerts, M.; Everaert, H.; et al. Undetectable circulating tumor DNA (ctDNA) levels correlate with favorable outcome in metastatic melanoma patients treated with anti-PD1 therapy. J. Transl. Med. 2019, 17, 303. [Google Scholar] [CrossRef]
- Ma, C.; Yang, X.; Xing, W.; Yu, H.; Si, T.; Guo, Z. Detection of circulating tumor DNA from non-small cell lung cancer brain metastasis in cerebrospinal fluid samples. Thorac. Cancer 2020, 11, 588–593. [Google Scholar] [CrossRef]
- Sato, J.; Shimomura, A.; Kawauchi, J.; Matsuzaki, J.; Yamamoto, Y.; Takizawa, S.; Sakamoto, H.; Ohno, M.; Narita, Y.; Ochiya, T.; et al. Brain metastasis-related microRNAs in patients with advanced breast cancer. PLoS ONE 2019, 14, e0221538. [Google Scholar] [CrossRef] [PubMed]
- Bustos, M.A.; Tran, K.D.; Rahimzadeh, N.; Gross, R.; Lin, S.Y.; Shoji, Y.; Murakami, T.; Boley, C.L.; Tran, L.T.; Cole, H.; et al. Integrated Assessment of Circulating Cell-Free MicroRNA Signatures in Plasma of Patients with Melanoma Brain Metastasis. Cancers 2020, 12, 1692. [Google Scholar] [CrossRef]
- Teplyuk, N.M.; Mollenhauer, B.; Gabriely, G.; Giese, A.; Kim, E.; Smolsky, M.; Kim, R.Y.; Saria, M.G.; Pastorino, S.; Kesari, S.; et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro-Oncology 2012, 14, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Debeb, B.G.; Lacerda, L.; Anfossi, S.; Diagaradjane, P.; Chu, K.; Bambhroliya, A.; Huo, L.; Wei, C.; Larson, R.A.; Wolfe, A.R.; et al. miR-141-Mediated Regulation of Brain Metastasis From Breast Cancer. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef]
- Maji, S.; Chaudhary, P.; Akopova, I.; Nguyen, P.M.; Hare, R.J.; Gryczynski, I.; Vishwanatha, J.K. Exosomal Annexin II Promotes Angiogenesis and Breast Cancer Metastasis. Mol. Cancer Res. 2017, 15, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Morad, G.; Carman, C.V.; Hagedorn, E.J.; Perlin, J.R.; Zon, L.I.; Mustafaoglu, N.; Park, T.E.; Ingber, D.E.; Daisy, C.C.; Moses, M.A. Tumor-Derived Extracellular Vesicles Breach the Intact Blood-Brain Barrier via Transcytosis. ACS Nano 2019, 13, 13853–13865. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.-C.; Li, P.; Li, M.; Wang, X.; Zhang, C. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef]
- Wang, S.; Liang, K.; Hu, Q.; Li, P.; Song, J.; Yang, Y.; Yao, J.; Mangala, L.S.; Li, C.; Yang, W. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J. Clin. Investig. 2017, 127, 4498–4515. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Endo, H.; Yokoyama, M.; Abe, J.; Tamai, K.; Tanaka, N.; Sato, I.; Takahashi, S.; Kondo, T.; Satoh, K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2013, 436, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Chen, L.; Wang, Y.; Jiang, X.; Xia, H.; Zhuang, Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J. Neuro-Oncol. 2015, 121, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Fu, F.; Zhang, Y.; Gao, Z.; Zhao, Y.; Wen, Z.; Han, H.; Li, Y.; Chen, H. Development and validation of a five-gene model to predict postoperative brain metastasis in operable lung adenocarcinoma. Int. J. Cancer 2020, 147, 584–592. [Google Scholar] [CrossRef]
- Mueller, W.C.; Spector, Y.; Edmonston, T.B.; St Cyr, B.; Jaeger, D.; Lass, U.; Aharonov, R.; Rosenwald, S.; Chajut, A. Accurate classification of metastatic brain tumors using a novel microRNA-based test. Oncologist 2011, 16, 165–174. [Google Scholar] [CrossRef]
- Barciszewska, A.M. Global DNA demethylation as an epigenetic marker of human brain metastases. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Salomon, M.P.; Orozco, J.I.J.; Wilmott, J.S.; Hothi, P.; Manughian-Peter, A.O.; Cobbs, C.S.; Scolyer, R.A.; Hoon, D.S.B.; Marzese, D.M. Brain metastasis DNA methylomes, a novel resource for the identification of biological and clinical features. Sci. Data 2018, 5, 180245. [Google Scholar] [CrossRef]
- Marzese, D.M.; Scolyer, R.A.; Roque, M.; Vargas-Roig, L.M.; Huynh, J.L.; Wilmott, J.S.; Murali, R.; Buckland, M.E.; Barkhoudarian, G.; Thompson, J.F.; et al. DNA methylation and gene deletion analysis of brain metastases in melanoma patients identifies mutually exclusive molecular alterations. Neuro-Oncology 2014, 16, 1499–1509. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.D.; Johnson, M.D.; Ahmed, S.; Cardenas, P.Y.; Grills, I.S.; Thibodeau, B.J. Targeted DNA sequencing of non-small cell lung cancer identifies mutations associated with brain metastases. Oncotarget 2018, 9, 25957–25970. [Google Scholar] [CrossRef] [PubMed]
- Winther-Larsen, A.; Hviid, C.V.B.; Meldgaard, P.; Sorensen, B.S.; Sandfeld-Paulsen, B. Neurofilament Light Chain as A Biomarker for Brain Metastases. Cancers 2020, 12, 2852. [Google Scholar] [CrossRef]
- Rodrigues, G.; Hoshino, A.; Kenific, C.M.; Matei, I.R.; Steiner, L.; Freitas, D.; Kim, H.S.; Oxley, P.R.; Scandariato, I.; Casanova-Salas, I. Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol. 2019, 21, 1403–1412. [Google Scholar] [CrossRef]
- Han, L.; Liang, X.H.; Chen, L.X.; Bao, S.M.; Yan, Z.Q. SIRT1 is highly expressed in brain metastasis tissues of non-small cell lung cancer (NSCLC) and in positive regulation of NSCLC cell migration. Int. J. Clin. Exp. Pathol. 2013, 6, 2357–2365. [Google Scholar] [PubMed]
- Gril, B.; Wei, D.; Zimmer, A.S.; Robinson, C.; Khan, I.; Difilippantonio, S.; Overstreet, M.G.; Steeg, P.S. HER2 antibody-drug conjugate controls growth of breast cancer brain metastases in hematogenous xenograft models, with heterogeneous blood-tumor barrier penetration unlinked to a passive marker. Neuro-Oncology 2020, 22, 1625–1636. [Google Scholar] [CrossRef]
- Kuo, A.H.; Clarke, M.F. Identifying the metastatic seeds of breast cancer. Nat. Biotechnol. 2013, 31, 504–505. [Google Scholar] [CrossRef]
- Rack, B.; Schindlbeck, C.; Jückstock, J.; Andergassen, U.; Hepp, P.; Zwingers, T.; Friedl, T.W.P.; Lorenz, R.; Tesch, H.; Fasching, P.A.; et al. Circulating Tumor Cells Predict Survival in Early Average-to-High Risk Breast Cancer Patients. JNCI J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [PubMed]
- Sidaway, P. Brain metastasis detectable in CTCs. Nat. Rev. Clin. Oncol. 2017, 14, 588. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Pantel, K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer 2019, 19, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, A.; Riebensahm, C.; Mohme, M.; Joosse, S.A.; Velthaus, J.-L.; Berger, L.A.; Bernreuther, C.; Glatzel, M.; Loges, S.; Lamszus, K.; et al. Frequency of Circulating Tumor Cells (CTC) in Patients with Brain Metastases: Implications as a Risk Assessment Marker in Oligo-Metastatic Disease. Cancers 2018, 10, 527. [Google Scholar] [CrossRef] [PubMed]
- Veridex, L. CellSearch circulating tumor cell kit premarket notification-expanded indications for use—Metastatic prostate cancer. J. Clin. Oncol. 2015, 33, 1348–1355. [Google Scholar]
- Mego, M.; De Giorgi, U.; Dawood, S.; Wang, X.; Valero, V.; Andreopoulou, E.; Handy, B.; Ueno, N.T.; Reuben, J.M.; Cristofanilli, M. Characterization of metastatic breast cancer patients with nondetectable circulating tumor cells. Int. J. Cancer 2011, 129, 417–423. [Google Scholar] [CrossRef]
- Huebner, H.; Fasching, P.A.; Gumbrecht, W.; Jud, S.; Rauh, C.; Matzas, M.; Paulicka, P.; Friedrich, K.; Lux, M.P.; Volz, B.; et al. Filtration based assessment of CTCs and CellSearch® based assessment are both powerful predictors of prognosis for metastatic breast cancer patients. BMC Cancer 2018, 18, 204. [Google Scholar] [CrossRef]
- Deng, Y.; Zhang, Y.; Sun, S.; Wang, Z.; Wang, M.; Yu, B.; Czajkowsky, D.M.; Liu, B.; Li, Y.; Wei, W.; et al. An Integrated Microfluidic Chip System for Single-Cell Secretion Profiling of Rare Circulating Tumor Cells. Sci. Rep. 2014, 4, 7499. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, M.A.; Kallergi, G.; Zafeiriou, Z.; Manouras, L.; Theodoropoulos, P.A.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Co-expression of putative stemness and epithelial-to-mesenchymal transition markers on single circulating tumour cells from patients with early and metastatic breast cancer. BMC Cancer 2014, 14, 651. [Google Scholar] [CrossRef] [PubMed]
- Ossandon, M.R.; Agrawal, L.; Bernhard, E.J.; Conley, B.A.; Dey, S.M.; Divi, R.L.; Guan, P.; Lively, T.G.; McKee, T.C.; Sorg, B.S.; et al. Circulating Tumor DNA Assays in Clinical Cancer Research. JNCI J. Natl. Cancer Inst. 2018, 110, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Connolly, I.D.; Li, Y.; Gephart, M.H.; Nagpal, S. The “liquid biopsy”: The role of circulating DNA and RNA in central nervous system tumors. Curr. Neurol. Neurosci. Rep. 2016, 16, 25. [Google Scholar] [CrossRef]
- Chang, G.A.; Tadepalli, J.S.; Shao, Y.; Zhang, Y.; Weiss, S.; Robinson, E.; Spittle, C.; Furtado, M.; Shelton, D.N.; Karlin-Neumann, G. Sensitivity of plasma BRAFmutant and NRASmutant cell—Free DNA assays to detect metastatic melanoma in patients with low RECIST scores and non-RECIST disease progression. Mol. Oncol. 2016, 10, 157–165. [Google Scholar] [CrossRef]
- Research, G.V. Exosomes Market Size to Reach $2.28 Billion by 2030|CAGR: 18.8%. Available online: https://www.grandviewresearch.com/press-release/global-exosomes-market (accessed on 1 August 2021).
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Mohammadi, S.; Yousefi, F.; Shabaninejad, Z.; Movahedpour, A.; Mahjoubin Tehran, M.; Shafiee, A.; Moradizarmehri, S.; Hajighadimi, S.; Savardashtaki, A.; Mirzaei, H. Exosomes and cancer: From oncogenic roles to therapeutic applications. IUBMB Life 2020, 72, 724–748. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.; Chin, A.R. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, N.; Kosaka, N.; Ono, M.; Katsuda, T.; Yoshioka, Y.; Tamura, K.; Lötvall, J.; Nakagama, H.; Ochiya, T. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood–brain barrier. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef]
- Ippen, F.M.; Colman, H.; van den Bent, M.J.; Brastianos, P.K. Precision Medicine for Primary Central Nervous System Tumors: Are We There Yet? In American Society of Clinical Oncology Educational Book; American Society of Clinical Oncology: Alexandria, VA, USA, 2018; pp. 158–167. [Google Scholar] [CrossRef]
- Kalita-de Croft, P.; Sadeghi Rad, H.; Gasper, H.; O’Byrne, K.; Lakhani, S.R.; Kulasinghe, A. Spatial profiling technologies and applications for brain cancers. Expert Rev. Mol. Diagn. 2021, 1–10. [Google Scholar] [CrossRef]
- Ghiaseddin, A.; Hoang Minh, L.B.; Janiszewska, M.; Shin, D.; Wick, W.; Mitchell, D.A.; Wen, P.Y.; Grossman, S.A. Adult precision medicine: Learning from the past to enhance the future. Neuro-Oncol. Adv. 2020, 3, vdaa145. [Google Scholar] [CrossRef] [PubMed]
- Berghoff, A.S.; Brastianos, P.K. Toward Precision Medicine in Brain Metastases. Semin. Neurol. 2018, 38, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef] [PubMed]
- Misyura, M.; Zhang, T.; Sukhai, M.A.; Thomas, M.; Garg, S.; Kamel-Reid, S.; Stockley, T.L. Comparison of Next-Generation Sequencing Panels and Platforms for Detection and Verification of Somatic Tumor Variants for Clinical Diagnostics. J. Mol. Diagn. 2016, 18, 842–850. [Google Scholar] [CrossRef]
- Kalita-de Croft, P.; Al-Ejeh, F.; McCart Reed, A.E.; Saunus, J.M.; Lakhani, S.R. ‘Omics Approaches in Breast Cancer Research and Clinical Practice. Adv. Anat. Pathol. 2016, 23, 356–367. [Google Scholar] [CrossRef]
- Brastianos, P.K.; Carter, S.L.; Santagata, S.; Cahill, D.P.; Taylor-Weiner, A.; Jones, R.T.; Van Allen, E.M.; Lawrence, M.S.; Horowitz, P.M.; Cibulskis, K.; et al. Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov. 2015, 5, 1164. [Google Scholar] [CrossRef] [PubMed]
- Saunus, J.M.; Quinn, M.C.; Patch, A.M.; Pearson, J.V.; Bailey, P.J.; Nones, K.; McCart Reed, A.E.; Miller, D.; Wilson, P.J.; Al-Ejeh, F.; et al. Integrated genomic and transcriptomic analysis of human brain metastases identifies alterations of potential clinical significance. J. Pathol. 2015, 237, 363–378. [Google Scholar] [CrossRef]
- Tyran, M.; Carbuccia, N.; Garnier, S.; Guille, A.; Adelaïde, J.; Finetti, P.; Touzlian, J.; Viens, P.; Tallet, A.; Goncalves, A. A comparison of DNA mutation and copy number profiles of primary breast cancers and paired brain metastases for identifying clinically relevant genetic alterations in brain metastases. Cancers 2019, 11, 665. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.; Im, S.-A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef]
- Tutt, A.N.J.; Garber, J.E.; Kaufman, B.; Viale, G.; Fumagalli, D.; Rastogi, P.; Gelber, R.D.; de Azambuja, E.; Fielding, A.; Balmaña, J.; et al. Adjuvant Olaparib for Patients with BRCA1- or BRCA2-Mutated Breast Cancer. N. Engl. J. Med. 2021, 384, 2394–2405. [Google Scholar] [CrossRef]
- Diossy, M.; Reiniger, L.; Sztupinszki, Z.; Krzystanek, M.; Timms, K.M.; Neff, C.; Solimeno, C.; Pruss, D.; Eklund, A.C.; Toth, E. Breast cancer brain metastases show increased levels of genomic aberration-based homologous recombination deficiency scores relative to their corresponding primary tumors. Ann. Oncol. 2018, 29, 1948–1954. [Google Scholar] [CrossRef]
- Sun, J.; Wang, C.; Zhang, Y.; Xu, L.; Fang, W.; Zhu, Y.; Zheng, Y.; Chen, X.; Xie, X.; Hu, X. Genomic signatures reveal DNA damage response deficiency in colorectal cancer brain metastases. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Da Silva, L.; Simpson, P.T.; Smart, C.E.; Cocciardi, S.; Waddell, N.; Lane, A.; Morrison, B.J.; Vargas, A.C.; Healey, S.; Beesley, J.; et al. HER3 and downstream pathways are involved in colonization of brain metastases from breast cancer. Breast Cancer Res. 2010, 12, R46. [Google Scholar] [CrossRef]
- Kalita-de Croft, P.; Lim, M.; Chittoory, H.; de Luca, X.M.; Kutasovic, J.R.; Day, B.W.; Al-Ejeh, F.; Simpson, P.T.; McCart Reed, A.E.; Lakhani, S.R. Clinicopathologic significance of nuclear HER4 and phospho-YAP (S127) in human breast cancers and matching brain metastases. Ther. Adv. Med. Oncol. 2020, 12, 1758835920946259. [Google Scholar] [CrossRef] [PubMed]
- Kodack, D.P.; Askoxylakis, V.; Ferraro, G.B.; Sheng, Q.; Badeaux, M.; Goel, S.; Qi, X.; Shankaraiah, R.; Cao, Z.A.; Ramjiawan, R.R. The brain microenvironment mediates resistance in luminal breast cancer to PI3K inhibition through HER3 activation. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Varešlija, D.; Priedigkeit, N.; Fagan, A.; Purcell, S.; Cosgrove, N.; O’Halloran, P.J.; Ward, E.; Cocchiglia, S.; Hartmaier, R.; Castro, C.A.; et al. Transcriptome Characterization of Matched Primary Breast and Brain Metastatic Tumors to Detect Novel Actionable Targets. JNCI J. Natl. Cancer Inst. 2019, 111, 388–398. [Google Scholar] [CrossRef]
- Gong, J.; Chehrazi-Raffle, A.; Reddi, S.; Salgia, R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: A comprehensive review of registration trials and future considerations. J. Immunother. Cancer 2018, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Akinleye, A.; Rasool, Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J. Hematol. Oncol. 2019, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Tawbi, H.A.; Chung, C.; Margolin, K. Nivolumab and Ipilimumab in Melanoma Metastatic to the Brain. N. Engl. J. Med. 2018, 379, 2178. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Rizvi, N.A.; Goldman, J.W.; Gettinger, S.N.; Borghaei, H.; Brahmer, J.R.; Ready, N.E.; Gerber, D.E.; Chow, L.Q.; Juergens, R.A.; et al. Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): Results of an open-label, phase 1, multicohort study. Lancet Oncol. 2017, 18, 31–41. [Google Scholar] [CrossRef]
- Stein, M.K.; Pandey, M.; Xiu, J.; Tae, H.; Swensen, J.; Mittal, S.; Brenner, A.J.; Korn, W.M.; Heimberger, A.B.; Martin, M.G. Tumor Mutational Burden Is Site Specific in Non–Small-Cell Lung Cancer and Is Highest in Lung Adenocarcinoma Brain Metastases. JCO Precis. Oncol. 2019, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- McGrail, D.J.; Pilie, P.G.; Rashid, N.U.; Voorwerk, L.; Slagter, M.; Kok, M.; Jonasch, E.; Khasraw, M.; Heimberger, A.B.; Lim, B.; et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann. Oncol. 2021, 32, 661–672. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).