Multigene Family of Pore-Forming Toxins from Sea Anemone Heteractis crispa
Abstract
:1. Introduction
2. Results and Discussion
2.1. Precursor Primary Structure
2.2. Actinoporins Pertain to a Multigene Family
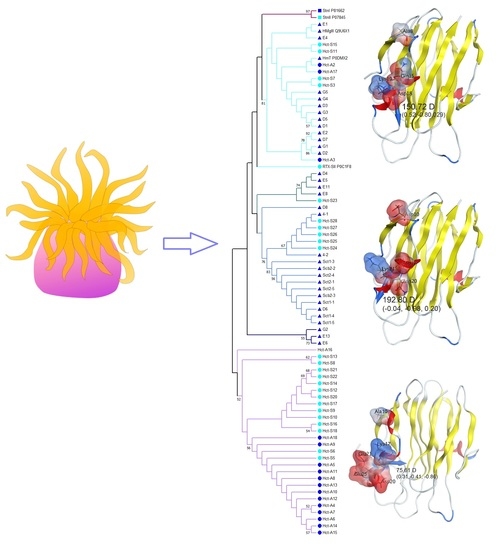
2.3. Phylogenetic and Sequences Analysis of Actinoporins
2.4. In Silico Analysis
2.5. Hemolytic Activity of the Recombinant Actinoporins
3. Materials and Methods
3.1. Precursor Determination
3.2. Cloning of cDNA Encoding Actinoporins
3.3. Recombinant Actinoporins Production
3.4. Sequences and Phylogenetic Analysis
3.5. Homology Modeling of Actinoporins
3.6. Hemolytic Assay
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Maček, P. Polypeptide cytolytic toxins from sea anemones (Actinaria). FEMS Microbiol. Immunol. 1992, 5, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Anderluh, G.; Maček, P. Cytolytic peptide and protein toxins from sea anemones (Anthozoa: Actiniaria). Toxicon 2002, 40, 111–124. [Google Scholar] [CrossRef]
- Parker, M.W.; Feil, S.C. Pore-forming protein toxins: From structure to function. Prog. Biophys. Mol. Biol. 2005, 88, 91–142. [Google Scholar] [CrossRef] [PubMed]
- García-Linares, S.; Rivera-de-Torre, E.; Palacios-Ortega, J.; Gavilanes, J.G.; Martínez-del-Pozo, Á. The metamorphic transformation of a water-soluble monomeric protein into an oligomeric transmembrane pore. In Advances in Biomembranes and Lipid Self-Assembly; Academic Press: Cambridge, MA, USA, 2017; Volume 26, pp. 51–97. [Google Scholar]
- Athanasiadis, A.; Anderluh, G.; Maček, P.; Turk, D. Crystal structure of the soluble form of equinatoxin II, a pore-forming toxin from the sea anemone Actinia equina. Structure 2001, 9, 341–346. [Google Scholar] [CrossRef]
- Gutiérrez-Aguirre, I.; Trontelj, P.; Maček, P.; Lakey, J.H.; Anderluh, G. Membrane binding of zebrafish actinoporin-like protein: AF domains, a novel superfamily of cell membrane binding domains. Biochem. J. 2006, 398, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Prentis, P.J.; Pavasovic, A.; Norton, R.S. Sea anemones: Quiet achievers in the field of peptide toxins. Toxins 2018, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Macrander, J.; Daly, M. Evolution of the cytolytic pore-forming proteins (Actinoporins) in sea anemones. Toxins 2016, 8, 368. [Google Scholar] [CrossRef] [PubMed]
- Maček, P.; Lebez, D. Kinetics of hemolysis induced by equinatoxin, a cytolytic toxin from the sea anemone Actinia equina. Effect of some ions and pH. Toxicon 1981, 19, 233–240. [Google Scholar] [CrossRef]
- Belmonte, G.; Menestrina, G.; Pederzolli, C.; Krizaj, I.; Gubensek, F.; Turk, T.; Macek, P. Primary and secondary structure of a pore-forming toxin from the sea anemone, Actinia equina L., and its association with lipid vesicles. Biochim. Biophys. Acta 1994, 1192, 197–204. [Google Scholar] [CrossRef]
- Anderluh, G.; Križaj, I.; Štrukelj, B.; Gubenšek, F.; Maček, P.; Pungerčar, J. Equinatoxins, pore-forming proteins from sea anemone Actinia equina, belong to a multigene family. Toxicon 1999, 37, 1391–1401. [Google Scholar] [CrossRef]
- Khoo, K.S.; Kam, W.K.; Khoo, H.E.; Gopalakrishnakone, P.; Chung, M.C.M. Purification and partial characterization of two cytolysins from a tropical sea anemone, Heteractis magnifica. Toxicon 1993, 31, 1567–1579. [Google Scholar] [CrossRef]
- Wang, Y.; Chua, K.L.; Khoo, H.E. A new cytolysin from the sea anemone Heteractis magnifica: Isolation cDNA cloning and functional expression. Biochem. Biophys. Acta 2000, 1478, 9–18. [Google Scholar] [CrossRef]
- Samejima, Y.; Yanagisaws, M.; Aoki-Tomomutsu, Y.; Iwasaki, E.; Ando, J.; Mebs, D. Amino acid sequence studies on cytolytic toxins from sea anemone Heteractis magnifica, Entacmaea quadricolor and Stichodactyla mertensii (Anthozoa). Toxicon 2000, 38, 259–264. [Google Scholar] [CrossRef]
- Lanio, M.E.; Morera, V.; Alvarez, C.; Tejuca, M.; Gomez, T.; Pazos, F.; Besada, V.; Martínez, D.; Huerta, V.; Padron, G.; et al. Purification and characterization of two hemolysins from Stichodactyla helianthus. Toxicon 2001, 39, 187–194. [Google Scholar] [CrossRef]
- Monastyrnaya, M.M.; Zykova, T.A.; Apalikova, O.V.; Shwets, T.V.; Kozlovskaya, E.P. Biologically active polypeptides from the tropical sea anemone Radianthus macrodactylus. Toxicon 2002, 40, 1197–1217. [Google Scholar] [CrossRef]
- Klyshko, E.V.; Issaeva, M.P.; Monastyrnaya, M.M.; Il’ina, A.P.; Guzev, K.V.; Vakorina, T.I.; Dmitrenok, P.S.; Zykova, T.A.; Kozlovskaya, E.P. Isolation, properties and partial amino acid sequence of a new actinoporin from the sea anemone Radianthus macrodactylus. Toxicon 2004, 44, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Il’ina, A.; Lipkin, A.; Barsova, E.; Issaeva, M.; Leychenko, E.; Guzev, K.; Monastyrnaya, M.; Lukyanov, S.; Kozlovskaya, E. Amino acid sequence of RTX-A’s isoform actinoporin from the sea anemone Radianthus macrodactylus. Toxicon 2006, 47, 517–520. [Google Scholar] [CrossRef] [PubMed]
- Il’ina, A.P.; Monastyrnaya, M.M.; Issaeva, M.P.; Guzev, K.V.; Rasskasov, V.A.; Kozlovskaya, E.P. Primary structure of actinoporins from the sea anemone Oulactis orientalis. Bioorg. Chem. (Russ.) 2005, 31, 357–362. [Google Scholar] [CrossRef]
- Monastyrnaya, M.; Leychenko, E.; Issaeva, M.; Likhatskaya, G.; Zelepuga, E.; Kostina, E.; Trifonov, E.; Nurminski, E.; Kozlovskaya, E. Actinoporins from the sea anemones, tropical Radianthus macrodactylus and northern Oulactis orientalis: Comparative analysis of structure-function relationships. Toxicon 2010, 56, 1299–1314. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Oshiro, N.; Takuwa-Kuroda, K.; Iwanaga, S.; Nozaki, M.; Nakajima, T.A. A new polypeptide toxin from the nematocyst venom of an Okinawan sea anemone Phyllodiscus semoni (Japanese name “unbachi-isoginchaku”). Biosci. Biotechnol. Biochem. 2002, 66, 2621–2625. [Google Scholar] [CrossRef] [PubMed]
- Uechi, G.; Toma, H.; Arakawa, T.; Sato, Y. Molecular characterization on the genome structure of hemolysin toxin isoforms isolated from sea anemone Actineria villosa and Phyllodiscus semoni. Toxicon 2010, 56, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Uechi, G.; Toma, H.; Arakawa, T.; Sato, Y. Molecular cloning and functional expression of hemolysin from the sea anemone Actineria villosa. Protein Expr. Purif. 2005, 40, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yap, L.L.; Chua, K.L.; Khoo, H.E. A multigene family of Heteractis magnificalysins (HMgs). Toxicon 2008, 51, 1374–1382. [Google Scholar] [CrossRef] [PubMed]
- Tkacheva, E.S.; Leychenko, E.V.; Monastyrnaya, M.M.; Issaeva, M.P.; Zelepuga, E.A.; Anastuk, S.D.; Dmitrenok, P.S.; Kozlovskaya, E.P. New actinoporins from sea anemone Heteractis crispa: Cloning and functional expression. Biochemistry 2011, 76, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Leichenko, E.V.; Monastirnaya, M.M.; Zelepuga, E.A.; Tkacheva, E.S.; Isaeva, M.P.; Likhatskaya, G.N.; Anastyuk, S.D.; Kozlovskaya, E.P. Hct-A is a new actinoporin family from the Heteractis crispa sea anemone. Acta Nat. 2014, 6, 89–98. [Google Scholar]
- Valle, A.; Alvarado-Mesen, J.; Lanio, M.E.; Alvarez, C.; Barbosa, J.A.R.G.; Pazos, I.F. The multigene families of actinoporins (part I): Isoforms and genetic structure. Toxicon 2015, 103, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Hinds, M.G.; Zhang, W.; Anderluh, G.; Hansen, P.E.; Norton, R.S. Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II: Implications for pore formation. J. Mol. Biol. 2002, 315, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- García-Linares, S.; Castrillo, I.; Bruix, M.; Menéndez, M.; Alegre-Cebollada, J.; Martínez-del-Pozo, Á.; Gavilanes, J.G. Three-dimensional structure of the actinoporin sticholysin I. Influence of long-distance effects on protein function. Arch. Biochem. Biophys. 2013, 532, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ortega, L.; Alegre-Cebollada, J.; García-Linares, S.; Bruix, M.; Martínez-del-Pozo, A.; Gavilanes, J.G. The behaviour of sea anemone actinoporins at the water-membrane interface. Biochim. Biophys. Acta 2011, 1808, 2275–2288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mancheño, J.M.; Martin-Benito, J.; Martínez, M.; Gavilanes, J.G.; Hermoso, J.A. Crystal and electron microscopy structures of Sticholysin II actinoporin reveal insights into the mechanism of membrane pore formation. Structure 2003, 11, 1–20. [Google Scholar] [CrossRef]
- Bellomio, A.; Morante, K.; Barlič, A.; Gutiérrez-Aguirre, I.; Viguera, A.R.; González-Mañas, J.M. Purification, cloning and characterization of fragaceatoxin C, a novel actinoporin from the sea anemone Actinia fragacea. Toxicon 2009, 54, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Mechaly, A.E.; Bellomio, A.; Gil-Carton, D.; Morante, K.; Valle, M.; González-Mañas, J.M.; Guérin, D.M. Structural insights into the oligomerization and architecture of eukaryotic membrane pore-forming toxins. Structure 2011, 19, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Caaveiro, J.M.M.; Morante, K.; González-Mañas, J.M.; Tsumoto, K. Structural basis for self-assembly of a cytolytic pore lined by protein and lipid. Nat. Commun. 2015, 6, 6337. [Google Scholar] [CrossRef] [PubMed]
- Hong, Q.; Gutiérrez-Aguirre, I.; Barlič, A.; Malovrh, P.; Kristan, K.; Podlesek, Z.; Maček, P.; Turk, D.; González-Mañas, J.M.; Lakey, J.H.; et al. Two-step membrane binding by equinatoxin II, a pore-forming toxin from the sea anemone, involves an exposed aromatic cluster and a flexible helix. J. Biol. Chem. 2002, 277, 41916–41924. [Google Scholar] [CrossRef] [PubMed]
- Malovrh, P.; Viero, G.; Dalla Serra, M.; Podlesek, Z.; Lakey, J.H.; Maček, P.; Menestrina, G.; Anderluh, G. A novel mechanism of pore formation. Membrane penetration by the N-terminal amphipathic region of equinatoxin. J. Biol. Chem. 2003, 278, 22678–22685. [Google Scholar] [CrossRef] [PubMed]
- Kristan, K.; Podlesek, Z.; Hojnik, V.; Gutiérrez-Aguirre, I.; Gunčar, G.; Turk, D.; Gonzalez-Mañas, J.M.; Lakey, J.H.; Maček, P.; Anderluh, G. Pore formation by equinatoxin, an eukaryotic pore-forming toxin, requires a flexible N-terminal region and a stable β-sandwich. J. Biol. Chem. 2004, 279, 46509–46517. [Google Scholar] [CrossRef] [PubMed]
- Alegre-Cebollada, J.; Oñaderra, M.; Gavilanes, J.G.; Martínez-del-Pozo, Á. Sea anemone actinoporins: The transition from a folded soluble state to a functionally active membrane-bound oligomeric pore. Curr. Protein Pept. Sci. 2007, 8, 558–572. [Google Scholar] [CrossRef] [PubMed]
- Bakrač, B.; Gutiérrez-Aguirre, I.; Podlesek, Z.; Sonnen, A.F.-P.; Gilbert, R.J.C.; Maček, P.; Lakey, J.H.; Anderluh, G. Molecular determinants of sphingomyelin specificity of a eukaryotic pore-forming toxin. J. Biol. Chem. 2008, 283, 18665–18677. [Google Scholar] [CrossRef] [PubMed]
- Kristan, K.; Vierob, G.; Dalla Serrab, M.; Maček, P.; Anderluh, G. Molecular mechanism of pore formation by actinoporins. Toxicon 2009, 54, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Bakrač, B.; Anderluh, G. Molecular mechanism of sphingomyelin-specific membrane binding and pore formation by actinoporins. Adv. Exp. Med. Biol. 2010, 677, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Anderluh, G.; Pungercar, J.; Krizaj, I.; Strukelj, B.; Gubensek, F.; Maček, P. N-terminal truncation mutagenesis of equinatoxin II, a pore-forming protein from the sea anemone Actinia equina. Protein Eng. 1997, 10, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Anderluh, G.; Barlic, A.; Krizaj, I.; Menestrina, G.; Gubensek, F.; Macek, P. Avidin-FITC topological studies with three cysteine mutants of equinatoxin II, a sea anemone pore-forming protein. Biochem. Biophys. Res. Commun. 1998, 242, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Aguirre, I.; Barlič, A.; Podlesek, Z.; Maček, P.; Anderluh, G.; González-Mañas, J.M. Membrane insertion of the N-terminal alpha-helix of equinatoxin II, a sea anemone cytolytic toxin. Biochem. J. 2004, 384, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Alegre-Cebollada, J.; Cunietti, M.; Herrero-Galán, E.; Gavilanes, J.G.; Martínez-del-Pozo, Á. Calorimetric scrutiny of lipid binding by sticholysin II toxin mutants. J. Mol. Biol. 2008, 382, 920–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malovrh, P.; Barlič, A.; Podlesek, Z.; Maček, P.; Menestrina, G.; Anderluh, G. Structure-function studies of tryptophan mutants of equinatoxin II, a sea anemone pore-forming protein. Biochem. J. 2000, 346, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Rojko, N.; Kristan, K.C.; Viero, G.; Žerovnik, E.; Maček, P.; Dalla-Serra, M.; Anderluh, G. Membrane damage by an α-helical pore-forming protein, equinatoxin II, proceeds through a succession of ordered steps. J. Biol. Chem. 2013, 288, 23704–23715. [Google Scholar] [CrossRef] [PubMed]
- Antonini, V.; Perez-Barzaga, V.; Bampi, S.; Pentón, D.; Martínez, D.; Dalla-Serra, M.; Tejuca, M. Functional characterization of sticholysin I and W111C mutant reveals the sequence of the actinoporin’s pore assembly. PLoS ONE 2014, 9, e110824. [Google Scholar] [CrossRef] [PubMed]
- Morante, K.; Caaveiro, J.M.M.; Viguera, A.R.; Tsumoto, K.; González-Mañas, J.M. Functional characterization of Val60, a key residue involved in the membrane oligomerization of fragaceatoxin C, an actinoporin from Actinia fragacea. FEBS Lett. 2015, 589, 1840–1846. [Google Scholar] [CrossRef] [PubMed]
- García-Linares, S.; Alm, I.; Maula, T.; Gavilanes, J.G.; Slotte, J.P.; Martínez-del-Pozo, Á. The effect of cholesterol on the long-range network of interactions established among sea anemone sticholysin II residues at the water-membrane interface. Mar. Drugs 2015, 13, 1647–1665. [Google Scholar] [CrossRef] [PubMed]
- Morante, K.; Caaveiro, J.M.M.; Tanaka, K.; González-Mañas, J.M.; Tsumoto, K. A pore-forming toxin requires a specific residue for its activity in membranes with particular physicochemical properties. J. Biol. Chem. 2015, 290, 10850–10861. [Google Scholar] [CrossRef] [PubMed]
- García-Linares, S.; Maula, T.; Rivera-de-Torre, E.; Gavilanes, J.G.; Slotte, J.P.; Martínez-del-Pozo, Á. Role of the tryptophan residues in the specific interaction of the sea anemone Stichodactyla helianthus’s actinoporin sticholysin II with biological membranes. Biochemistry 2016, 55, 6406–6420. [Google Scholar] [CrossRef] [PubMed]
- Mesa-Galloso, H.; Delgado-Magnero, K.H.; Cabezas, S.; López-Castilla, A.; Hernández-González, J.E.; Pedrera, L.; Alvarez, C.; Peter Tieleman, D.; García-Sáez, A.J.; Lanio, M.E.; et al. Disrupting a key hydrophobic pair in the oligomerization interface of the actinoporins impairs their pore-forming activity. Protein Sci. 2017, 26, 550–565. [Google Scholar] [CrossRef] [PubMed]
- Anderluh, G.; Lakey, J.H. Disparate proteins use similar architectures to damage membranes. Trends Biochem. Sci. 2008, 33, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Peterzolli, C.; Belmonte, G.; Dalla Serra, M.; Maček, P.; Menestrina, G. Biochemical and Cytotoxic Properties of Conjugates of Transferrin with Equinatoxin II, a Cytolysin from a Sea Anemone. Bioconjug. Chem. 1995, 6, 166–173. [Google Scholar] [CrossRef]
- Tejuca, M.; Anderluh, G.; Maček, P.; Marcet, R.; Torres, D.; Sarracent, J.; Alvarez, C.; Lanio, M.E.; Serra, M.D.; Menestrina, G. Antiparasite activity of sea-anemone cytolysins on Giardia duodenalis and specific targeting with anti-Giardia antibodies. Int. J. Parasitol. 1999, 29, 489–498. [Google Scholar] [CrossRef]
- Tejuca, M.; Anderluh, G.; Dalla Serra, M. Sea anemone cytolysins as toxic components of immunotoxins. Toxicon 2009, 54, 1206–1214. [Google Scholar] [CrossRef] [PubMed]
- Laborde, R.J.; Sanchez-Ferras, O.; Luzardo, M.C.; Cruz-Leal, Y.; Fernández, A.; Mesa, C.; Oliver, L.; Canet, L.; Abreu-Butin, L.; Nogueira, C.V.; et al. Novel adjuvant based on the pore-forming protein Sticholysin II encapsulated into liposomes effectively enhances the antigen-specific CTL-mediated immune response. J. Immunol. 2017, 198, 2772–2784. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, S.; Dyshlovoy, S.; Monastyrnaya, M.; Shubina, L.; Leychenko, E.; Kozlovskaya, E.; Jin, J.O.; Kwak, J.Y.; Bode, A.M.; Dong, Z.; et al. The anticancer effects of actinoporin RTX-A from the sea anemone Heteractis crispa (=Radianthus macrodactylus). Toxicon 2010, 55, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Matz, M.V.; Alieva, N.O.; Chenchik, A.; Lukyanov, S. Amplification of cDNA ends using PCR suppression effect and step-out PCR. Methods Mol. Biol. 2003, 221, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. An analysis of 5′-noncoding sequences from 669 vertebrate messenger RNAs. Nucleic Acids Res. 1987, 15, 8125–8148. [Google Scholar] [CrossRef] [PubMed]
- Von Heijne, G. Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biohem. 1983, 133, 17–21. [Google Scholar] [CrossRef]
- Pungerčar, J.; Anderluh, G.; Gubenšek, F.; Štrukelj, B. Sequence analysis of the cDNA encoding the precursor of equinatoxin V, a newly discovered hemolysin from the sea anemone Actinia equina. Biochem. Biophys. Acta 1997, 1341, 105–107. [Google Scholar] [CrossRef]
- Anderluh, G.; Podlesek, Z.; Maček, P. A common motif in proparts of Cnidarian toxins and nematocyst collagens and its putative role. Biochim. Biophys. Acta 2000, 1476, 372–376. [Google Scholar] [CrossRef]
- Olivera, B.M.; Walker, C.; Cartier, G.E.; Hooper, D.; Santos, A.D.; Schoenfeld, R.; Shetty, R.; Watkins, M.; Bandyopadhyay, P.; Hillyard, D.R. Speciation of cone snails and interspecific hyperdivergence of their venom peptides. Potential evolutionary significance of introns. Ann. N. Y. Acad. Sci. 1999, 870, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Pineda, S.S.; Sollod, B.L.; Wilson, D.; Darling, A.; Sunagar, K.; Undheim, E.A.; Kely, L.; Antunes, A.; Fry, B.G.; King, G.F. Diversification of a single ancestral gene into a successful toxin superfamily in highly venomous Australian funnel-web spiders. BMC Genom. 2014, 15, 177. [Google Scholar] [CrossRef] [PubMed]
- Leoni, G.; De Poli, A.; Mardirossian, M.; Gambato, S.; Florian, F.; Venier, P.; Wilson, D.N.; Tossi, A.; Pallavicini, A.; Gerdol, M. Myticalins: A novel multigenic family of linear, cationic antimicrobial peptides from marine mussels (Mytilus spp.). Mar. Drugs 2017, 15, 261. [Google Scholar] [CrossRef] [PubMed]
- Columbus-Shenkar, Y.Y.; Sachkova, M.Y.; Macrander, J.; Fridrich, A.; Modepalli, V.; Reitzel, A.M.; Sunagar, K.; Moran, Y. Dynamics of venom composition across a complex life cycle. Elife 2018, 7, e35014. [Google Scholar] [CrossRef] [PubMed]
- De los Ríos, V.; Oñaderra, M.; Martínez-Ruiz, A.; Lacadena, J.; Mancheño, J.M.; Martínez-Del-Pozo, A.; Gavilanes, J.G. Overproduction in Escherichia coli and purification of the hemolytic protein Sticholysin II from the sea anemone Stichodactyla helianthus. Protein Expr. Purif. 2000, 18, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Madio, B.; Undheim, E.A.B.; King, G.F. Revisiting venom of the sea anemone Stichodactyla haddoni: Omics techniques reveal the complete toxin arsenal of a well-studied sea anemone genus. J. Proteom. 2017, 166, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ari, H.; Paz, M.; Sher, D. The chemical armament of reef-building corals: Inter-and intra-specific variation and the identification of an unusual actinoporin in Stylophora pistilata. Sci. Rep. 2018, 8, 251. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, W.; Wang, L.; Wang, J.; Liu, X.; Jiao, B. Purification and characterization of gigantoxin-4, a new actinoporin from the sea anemone Stichodactyla gigantea. Int. J. Biol. Sci. 2011, 7, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Jouiaei, M.; Sunagar, K.; Federman Gross, A.; Scheib, H.; Alewood, P.F.; Moran, Y.; Fry, B.G. Evolution of an ancient venom: Recognition of a novel family of cnidarian toxins and the common evolutionary origin of sodium and potassium neurotoxins in sea anemone. Mol. Biol. Evol. 2015, 32, 1598–1610. [Google Scholar] [CrossRef] [PubMed]
- Ros, U.; Rodríguez-Vera, W.; Pedrera, L.; Valiente, P.A.; Cabezas, S.; Lanio, M.E.; García-Sáez, A.J.; Alvarez, C. Differences in activity of actinoporins are related with the hydrophobicity of their N-terminus. Biochimie 2015, 116, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Chemical Computing Group Inc. Molecular Operating Environment (MOE) 2016.08; Chemical Computing Group Inc.: Montreal, QC, Canada, 2016. [Google Scholar]
- Richter, S.; Wenzel, A.; Stein, M.; Gabdoulline, R.R.; Wade, R.C. webPIPSA: A web server for the comparison of protein interaction properties. Nucleic Acid Res. 2008, 36, W276–W280. [Google Scholar] [CrossRef] [PubMed]
- Rivera-de-Torre, E.; Palacios-Ortega, J.; García-Linares, S.; Gavilanes, J.G.; Martínez-Del-Pozo, Á. One single salt bridge explains the different cytolytic activities shown by actinoporins sticholysin I and II from the venom of Stichodactyla helianthus. Arch. Biochem. Biophys. 2017, 636, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Rivera-de-Torre, E.; García-Linares, S.; Alegre-Cebollada, J.; Lacadena, J.; Gavilanes, J.G.; Martínez-Del-Pozo, Á. Synergistic action of actinoporin isoforms from the same sea anemone species assembled into functionally active heteropores. J. Biol. Chem. 2016, 291, 14109–14119. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Eswar, N.; Marti-Renom, M.A.; Webb, B.; Madhusudhan, M.S.; Eramian, D.; Shen, M.; Pieper, U.; Sali, A. Comparative protein structure modeling with MODELLER. In Current Protocols in Bioinformatics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006; pp. 5.6:1–5.6:30. [Google Scholar]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK—A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]






| Actinoporin | Dipole Moment, Debye | Dipole Moment Direction, x, y, z |
|---|---|---|
| Hct-A2 | 150.72 | 0.52, −0.80, 0.29 |
| Hct-S3 | 132.89 | 0.11, −0.82, 0.56 |
| Hct-A17 | 102.90 | 0.45, −0.75, 0.48 |
| Hct-S17 | 134.30 | 0.48, −0.72, 0.21 |
| Hct-A6 | 69.66 | 0.82, −0.51, −0.07 |
| Hct-S5 | 75.61 | 0.31, −0.41, −0.86 |
| Hct-S23 | 192.80 | −0.04, −0.98, 0.20 |
| Hct-S25 | 159.07 | −0.06, −0.91, 0.41 |
| Sea anemone | Actinoporin | Molecular Mass, kDa | pI | Hemolytic Activity, HU/mg | Reference |
|---|---|---|---|---|---|
| H. crispa | rHct-S3 | 19.39 | 9.31 | (3.3 ± 0.31) × 104 | |
| rHct-A3 | 19.20 | 9.75 | (2.0 ± 0.49) × 104 | ||
| rHct-A5 | 19.21 | 9.45 | (1.0 ± 0.26) × 103 | ||
| rHct-A2 | 19.14 | 9.76 | (4.0 ± 0.36) × 104 | [26] | |
| rHct-S5 | 19.37 | 9.33 | (1.0 ± 0.53) × 103 | [25] | |
| rHct-S6 | 19.39 | 9.10 | (4.2 ± 0.52) × 103 | [25] | |
| RTX-S | ~20.00 | ~9.8 | 5.0 × 104 | [16] | |
| RTX-A | ~20.00 | ~9.8 | 3.5 × 104 | [16] | |
| RTX-G | ~20.00 | ~10.5 | 1.0 × 104 | [16] | |
| RTX-S II | 19.28 | 10 | 3.6 × 104 | [17] | |
| H. magnifica | HMgI | 19.0 | 9.4 | 3.6 × 104 | [12] |
| HMgII | 19.0 | 10.0 | 3.3 × 104 | [12] | |
| A. equina | EqtII | 19.0 | 10.5 | 3.6 × 104 | [9] |
| S. helianthus | StnI | 19.39 | 9 | 2.7 × 104 | [15] |
| StnII | 19.28 | 9 | 3.0 × 104 | [15] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leychenko, E.; Isaeva, M.; Tkacheva, E.; Zelepuga, E.; Kvetkina, A.; Guzev, K.; Monastyrnaya, M.; Kozlovskaya, E. Multigene Family of Pore-Forming Toxins from Sea Anemone Heteractis crispa. Mar. Drugs 2018, 16, 183. https://doi.org/10.3390/md16060183
Leychenko E, Isaeva M, Tkacheva E, Zelepuga E, Kvetkina A, Guzev K, Monastyrnaya M, Kozlovskaya E. Multigene Family of Pore-Forming Toxins from Sea Anemone Heteractis crispa. Marine Drugs. 2018; 16(6):183. https://doi.org/10.3390/md16060183
Chicago/Turabian StyleLeychenko, Elena, Marina Isaeva, Ekaterina Tkacheva, Elena Zelepuga, Aleksandra Kvetkina, Konstantin Guzev, Margarita Monastyrnaya, and Emma Kozlovskaya. 2018. "Multigene Family of Pore-Forming Toxins from Sea Anemone Heteractis crispa" Marine Drugs 16, no. 6: 183. https://doi.org/10.3390/md16060183
APA StyleLeychenko, E., Isaeva, M., Tkacheva, E., Zelepuga, E., Kvetkina, A., Guzev, K., Monastyrnaya, M., & Kozlovskaya, E. (2018). Multigene Family of Pore-Forming Toxins from Sea Anemone Heteractis crispa. Marine Drugs, 16(6), 183. https://doi.org/10.3390/md16060183