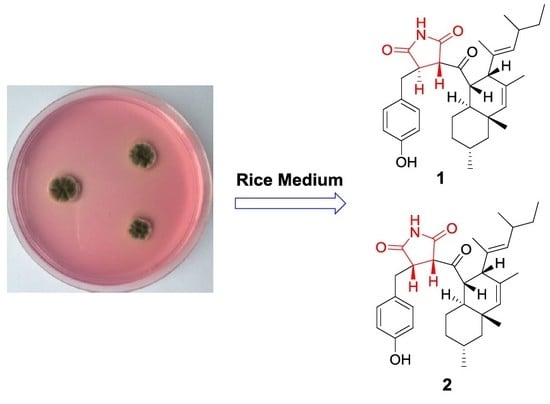
Two New Succinimide Derivatives Cladosporitins A and B from the Mangrove-derived Fungus Cladosporium sp. HNWSW-1
Abstract
:1. Introduction
2. Results
2.1. Structural Elucidation
2.2. The Bioactivities of Compounds 1–5 from Cladosporium sp. HNWSW-1
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation and Extraction
3.4. Purification and Identification
3.5. Bioassays for Cytotoxic Activity
3.6. Bioassays for α-Glycosidase Inhibitory Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2016, 33, 382–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.C.; Chen, C.M.; Tong, Q.Y.; Yang, J.; Wei, G.Z.; Xue, Y.B.; Wang, J.P.; Luo, Z.W.; Zhang, Y.H. Asperflavipine A: A cytochalasan heterotetramer uniquely defined by a highly complex tetradecacyclic ting dystem from Aspergillus flavipes QCS12. Angew. Chem. Int. Ed. 2017, 56, 5242–5246. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Wei, Y.J.; Ge, H.M.; Jiao, R.H.; Tan, R.X. Acaulide, an osteogenic macrodiolide from Acaulium sp. H-JQSF, an isopod-associated Fungus. Org. Lett. 2018, 20, 1007–1010. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Xu, R.; Li, X.M.; Yang, S.Q.; Meng, L.H.; Wang, B.G. Simpterpenoid A, a meroterpenoid with a highly functionalized cyclohexadiene moiety featuring gem-propane-1,2-dione and methylformate groups, from the mangrove-derived Penicillium simplicissimum MA-332. Org. Lett. 2018, 20, 1465–1468. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.D.; Ma, Q.Y.; Huang, S.Z.; Wang, P.; Wang, J.F.; Zhou, L.M.; Yuan, J.Z.; Dai, H.F.; Zhao, Y.X. Chrodrimanins K-N and related meroterpenoids from the fungus Penicillium sp. SCS-KFD09 isolated from a marine worm, Sipunculus nudus. J. Nat. Prod. 2017, 80, 1039–1047. [Google Scholar] [CrossRef]
- Suzuki, S.; Hosoe, T.; Nozawa, K.; Kawai, K.I.; Yaguchi, T.; Udagawa, S.I. Antifungal substances against pathogenic fungi, talaroconvolutins, from Talaromyces convolutes. J. Nat. Prod. 2000, 63, 768–772. [Google Scholar] [CrossRef]
- West, P.R.; van Ness, J.; Varming, A.-M.; Rassing, B.; Biggs, S.; Gasper, S.; McKernan, P.A.; Piggott, J. ZG- 1494α, a novel platelet-activating factor acetyltransferase inhibitor from Penicilium rubrum, isolation, structure elucidation and biological activity. J. Antibiot. 1996, 49, 967–973. [Google Scholar] [CrossRef]
- Sing, S.B.; Goetz, M.A.; Jones, T.; Bill, G.F.; Giacobbe, R.A.; Herram, L.; Mile, S.S.; Williams, D.L. Oteromycin: A novel antagonist of endothelin receptor. J.Org. Chem. 1995, 60, 7040–7042. [Google Scholar] [CrossRef]
- Kontik, R.; Clardy, J. Codinaeopsin, an antimalarial fungal polyketide. Org. Lett. 2008, 10, 4149–4151. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.P.; Wang, Y.; Zhu, G.L.; Zhu, W.M. Fatty acid derivatives from the halotolerant fungus Cladosporium cladosporioides. Magn. Reson. Chem. 2018, 56, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Jadulco, R.; Proksch, P.; Wray, V.; Albrecht Berg, S.; Grafe, U. New macrolides and furan carboxylic acid derivative from the sponge-derived fungus Cladosporium herbarum. J. Nat. Prod. 2001, 64, 527–530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tomoda, H.; Tabata, N.; Miura, H.; Namikoshi, M.; Yamaguchi, Y.; Masuma, R.; Omura, S. Cladospolide D, a new 12-membered macrolide antibiotic produced by Cladosporium sp. FT-0012. J. Antibiot. 2001, 54, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Shigemori, H.; Kasai, Y.; Komatsu, K.; Tsuda, M.; Mikami, Y.; Kobayashi, J.I. Sporiolides A and B, new cytotoxic twelve-membered macrolides from a marine-derived fungus Cladosporium species. Mar. Drugs 2004, 2, 164–169. [Google Scholar] [CrossRef]
- Jadulco, R.; Brauers, G.; Edrada, R.A.; Ebel, R.; Wray, V.; Sudarsono, S.; Proksch, P. New metabolites from sponge-derived fungi Curvularia lunata and Cladosporium herbarum. J. Nat. Prod. 2002, 65, 730–733. [Google Scholar] [CrossRef] [PubMed]
- Rotinsulu, H.; Yamazaki, H.; Sugai, S.; Iwakura, N.; Wewengkang, D.S.; Sumilat, D.A.; Namikoshi, M. Cladosporamide A, a new protein tyrosine phosphatase 1B inhibitor, produced by an Indonesian marine sponge-derived Cladosporium sp. J. Nat. Med. 2018. [Google Scholar] [CrossRef]
- Wang, L.P.; Han, X.L.; Zhu, G.L.; Wang, Y.; Chairoungdua, A.; Piyachaturawat, P.; Zhu, W.M. Polyketides from the endophytic fungus Cladosporium sp. isolated from the mangrove plant Excoecaria agallocha. Front. Chem. 2018, 6, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Sakagami, Y.; Sano, A.; Hara, O.; Mikawa, T.; Marumo, S. Cladosporol, β-1, 3-glucan biosynthesis inhibitor, isolated from fungus, Cladosporium cladosporioides. Tetrahedron Lett. 1995, 36, 1469–1472. [Google Scholar] [CrossRef]
- Yamazaki, H.; Yagi, A.; Akaishi, M.; Kirikoshi, R.; Takahashi, O.; Abe, T.; Chiba, S.; Takahashi, K.; Iwakura, N.; Namikoshi, M.; et al. Halogenated cladosporols produced by the sodium halide-supplemented fermentation of the plant-associated fungus Cladosporium sp. TMPU1621. Tetrahedron Lett. 2018, 59, 59,1913–1915. [Google Scholar] [CrossRef]
- Ye, Y.H.; Zhu, H.L.; Song, Y.C.; Liu, J.Y.; Tan, R.X. Structural revision of aspernigrin A, reisolated from Cladosporium herbarum IFB-E002. J. Nat. Prod. 2005, 68, 1106–1108. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Huang, Z.H.; Ma, X.; Qi, S.H. Unstable tetramic acid derivatives from the deep sea-derived fungus Cladosporium sphaerospermum EIODSF 008. Mar. Drugs 2018, 16, 448. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.L.; Kong, F.D.; Wang, Y.; Fu, P.; Zhu, W.M. Cladodionen, a cytotoxic hybrid polyketide from the marine-derived Cladosporium sp. OUCMDZ-1635. Mar. Drugs 2018, 16, 71. [Google Scholar] [CrossRef]
- Zhao, D.L.; Wang, D.; Tian, X.Y.; Cao, F.; Li, Y.Q.; Zhang, C.S. Anti-phytopathogenic and cytotoxic activities of crude extracts and secondary metabolites of marine-derived fungi. Mar. Drugs 2018, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, H.; Kamisuki, S.; Kaneko, M.; Yamaguguchi, Y.; Takeuchi, T.; Watashi, K.; Sugawara, F. Isolation and structure of vanitaracin A, a novel anti-hepatitis B virus compound from Talaromyces sp. Bioorg. Med. Chem. Lett. 2015, 25, 4325–4328. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Jong, A.N.; Bhandari, M.R.; Kawabata, J. α-Glucosidase inhibitors from Devil tree. Food Chem. 2007, 103, 1319–1323. [Google Scholar] [CrossRef]



| Position | 1 a | 2 b | ||
|---|---|---|---|---|
| δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, mult. (J in Hz) | |
| 1 | - | - | - | - |
| 2 | 172.7, C | - | 172.0, C | - |
| 3 | 61.7, CH | 4.06, d, (2.4) | 61.9, CH | 3.40, d, (4.1) |
| 4 | 45.0, CH | 3.40, m | 43.6, CH | 3.67, m |
| 5 | 179.2, C | 178.3, C | - | |
| 6 | 35.8, CH2 | 3.11, dd, (14.7, 5.9) 2.87, dd, (14.7, 5.3) | 35.0, CH2 | 3.10, dd, (13.7, 5.1) 2.79, dd, (13.7, 8.7) |
| 7 | 128.1, C | - | 128.2, C | - |
| 8 | 131.5, CH | 7.10, d, (8.3) | 130.5, CH | 6.99, d, (8.7) |
| 9 | 116.3, CH | 6.77, d, (8.3) | 115.9, CH | 6.75 d, (8.7) |
| 10 | 157.5, C | - | 155.4, C | - |
| 11 | 116.3, CH | 6.77, d (8.3) | 115.9, CH | 6.75, d, (8.7) |
| 12 | 131.5, CH | 7.10, d (8.3) | 130.5, CH | 6.99, d, (8.7) |
| 13 | 203.9, C | - | 202.2, C | - |
| 14 | 52.9, CH | 3.62, dd (12.0, 7.0) | 52.6, C | 3.33, dd, (12.1, 6.9) |
| 15 | 51.8, CH | 3.10 m | 50.9, CH | 2.95, d, (6.9) |
| 16 | 130.7, C | - | 130.1, C | - |
| 17 | 137.1, CH | 5.38, s | 135.8, CH | 5.29, s |
| 18 | 49.1, CH2 | 1.48, m 0.87, m | 48.3, CH2 | 1.43, m 0.86, m |
| 19 | 28.1, CH | 1.66, m | 27.4, CH | 1.61, m |
| 20 | 36.5, CH2 | 1.64, m 0.82, m | 35.6, CH2 | 1.60, m 0.77, m |
| 21 | 24.1, CH2 | 1.32, m 0.96, m | 24.3, CH2 | 1.62, m 0.75, m |
| 22 | 40.8, CH | 1.78, ddd, (12.0, 12.0, 2.3) | 42.2, CH | 1.67, m |
| 23 | 36.2, C | - | 35.2, C | - |
| 24 | 135.7, C | - | 135.0, C | - |
| 25 | 136.7, CH | 5.05, d, (8.2) | 136.1, CH | 4.66, d, (9.3) |
| 26 | 34.8, CH | 2.25, m | 33.9, CH | 2.14, m |
| 27 | 31.1, CH2 | 1.35, m 1.23, m | 30.4, CH2 | 1.27, m 1.10, m |
| 28 | 12.4, CH3 | 0.86, t, (7.6) | 12.1, CH3 | 0.79, t, (7.1) |
| 29 | 21.1, CH3 | 0.92, d, (6.2) | 20.9, CH3 | 0.76, d, (6.6) |
| 30 | 15.2, CH3 | 1.45, s | 14.5, CH3 | 1.47, s |
| 31 | 22.5, CH3 | 1.54, s | 22.2, CH3 | 1.46, s |
| 32 | 20.5, CH3 | 0.91, s | 20.3, CH3 | 0.85, s |
| 33 | 23.1, CH3 | 0.82, d, (6.4) | 22.8, CH3 | 0.82, d, (6.2) |
| Position | 3 | |
|---|---|---|
| δC, Type | δH, Mult. (J in Hz) | |
| 1 | - | - |
| 2 | 162.0, C | - |
| 3 | 109.9, CH | 6.01, s |
| 4 | 182.3, C | - |
| 4a | 115.8, C | - |
| 5 | 143.6, C | - |
| 6 | 118.3, CH | 6.64, dd, (2.2, 0.8) |
| 7 | 164.0, C | - |
| 8 | 101.8, CH | 6.70, d, (2.2) |
| 8a | 161.0, C | - |
| 1′ | 124.8, CH | 6.26, ddd, (15.6, 3.4, 1.7) |
| 2′ | 137.3, CH | 6.86, ddd, (15.6, 13.7, 6.9) |
| 3′ | 18.6, CH3 | 1.99, dd, (6.9,1.7) |
| 5- CH3 | 23.1, CH3 | 2.73, s |
| Compounds | IC50 (μM) | ||||
|---|---|---|---|---|---|
| Hela | BEL-7042 | K562 | SGC-7901 | α-Glycosidase | |
| 1 | >50 | >50 | >50 | >50 | >500 |
| 2 | >50 | 29.4 ± 0.35 | 25.6 ± 0.47 | 41.7 ± 0.71 | >500 |
| 3 | >50 | >50 | >50 | >50 | >500 |
| 4 | 14.9 ± 0.21 | 26.7 ± 1.1 | >50 | >50 | 78.2 ± 2.1 |
| 5 | >50 | >50 | >50 | >50 | 49.3 ± 10.6 |
| Adriamycin | 11.5 ± 0.18 | 11.9 ± 0.37 | 14.2 ± 0.66 | 6.66 ± 0.2 | ND a |
| Acarbose | ND a | ND a | ND a | ND a | 275.7 ± 2.7 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Cui, Y.; Cai, C.; Chen, H.; Dai, Y.; Chen, P.; Kong, F.; Yuan, J.; Song, X.; Mei, W.; et al. Two New Succinimide Derivatives Cladosporitins A and B from the Mangrove-derived Fungus Cladosporium sp. HNWSW-1. Mar. Drugs 2019, 17, 4. https://doi.org/10.3390/md17010004
Wang P, Cui Y, Cai C, Chen H, Dai Y, Chen P, Kong F, Yuan J, Song X, Mei W, et al. Two New Succinimide Derivatives Cladosporitins A and B from the Mangrove-derived Fungus Cladosporium sp. HNWSW-1. Marine Drugs. 2019; 17(1):4. https://doi.org/10.3390/md17010004
Chicago/Turabian StyleWang, Pei, Yan Cui, Caihong Cai, Huiqin Chen, Yu Dai, Pengwei Chen, Fandong Kong, Jingzhe Yuan, Xinming Song, Wenli Mei, and et al. 2019. "Two New Succinimide Derivatives Cladosporitins A and B from the Mangrove-derived Fungus Cladosporium sp. HNWSW-1" Marine Drugs 17, no. 1: 4. https://doi.org/10.3390/md17010004
APA StyleWang, P., Cui, Y., Cai, C., Chen, H., Dai, Y., Chen, P., Kong, F., Yuan, J., Song, X., Mei, W., & Dai, H. (2019). Two New Succinimide Derivatives Cladosporitins A and B from the Mangrove-derived Fungus Cladosporium sp. HNWSW-1. Marine Drugs, 17(1), 4. https://doi.org/10.3390/md17010004