Metabolomic and Transcriptomic Analyses of Escherichia coli for Efficient Fermentation of L-Fucose
Abstract
1. Introduction
2. Results
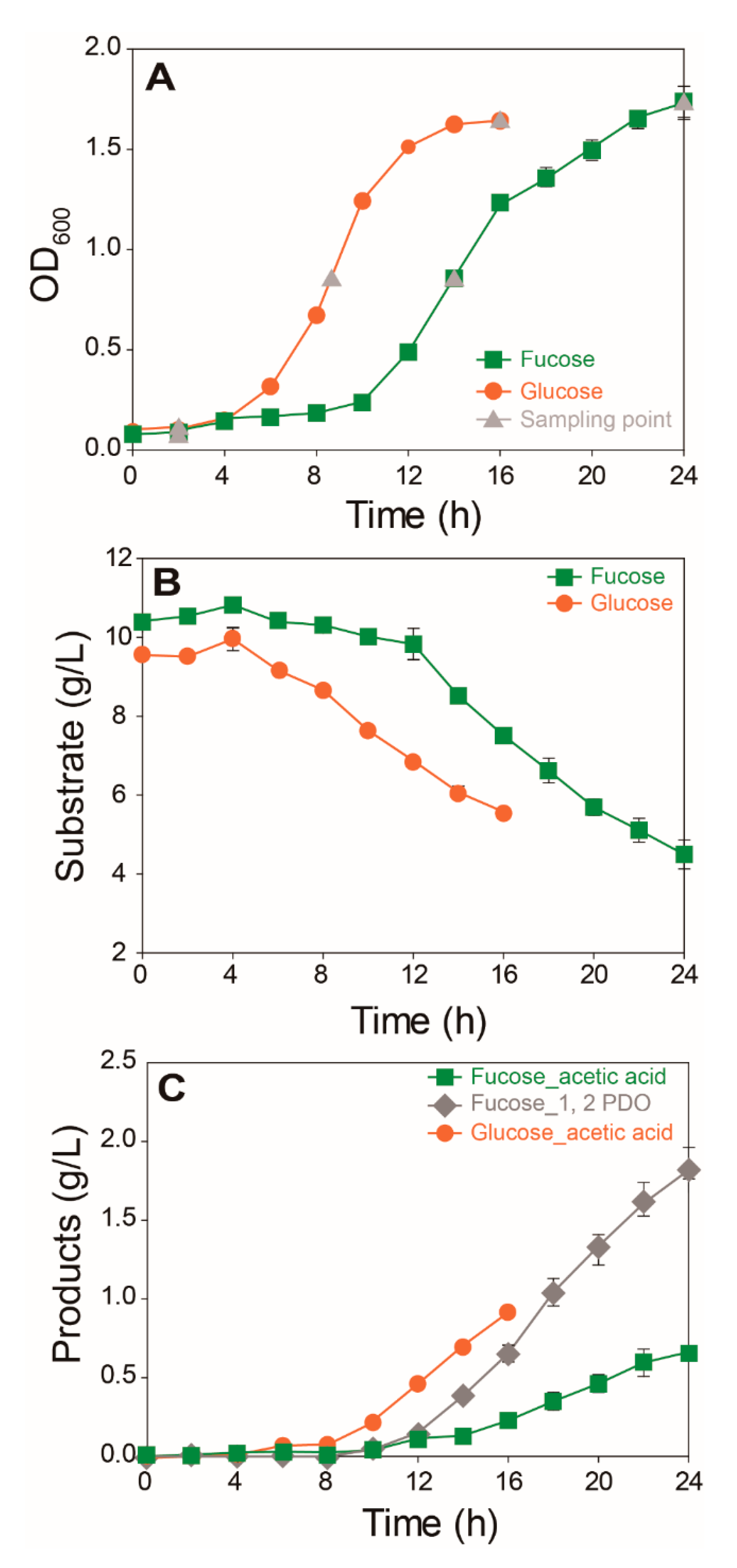
2.1. Comparison of Growth and Fermentation Product Profiles of E. coli on Fucose and Glucose
2.2. Identification of Metabolites and Transcripts
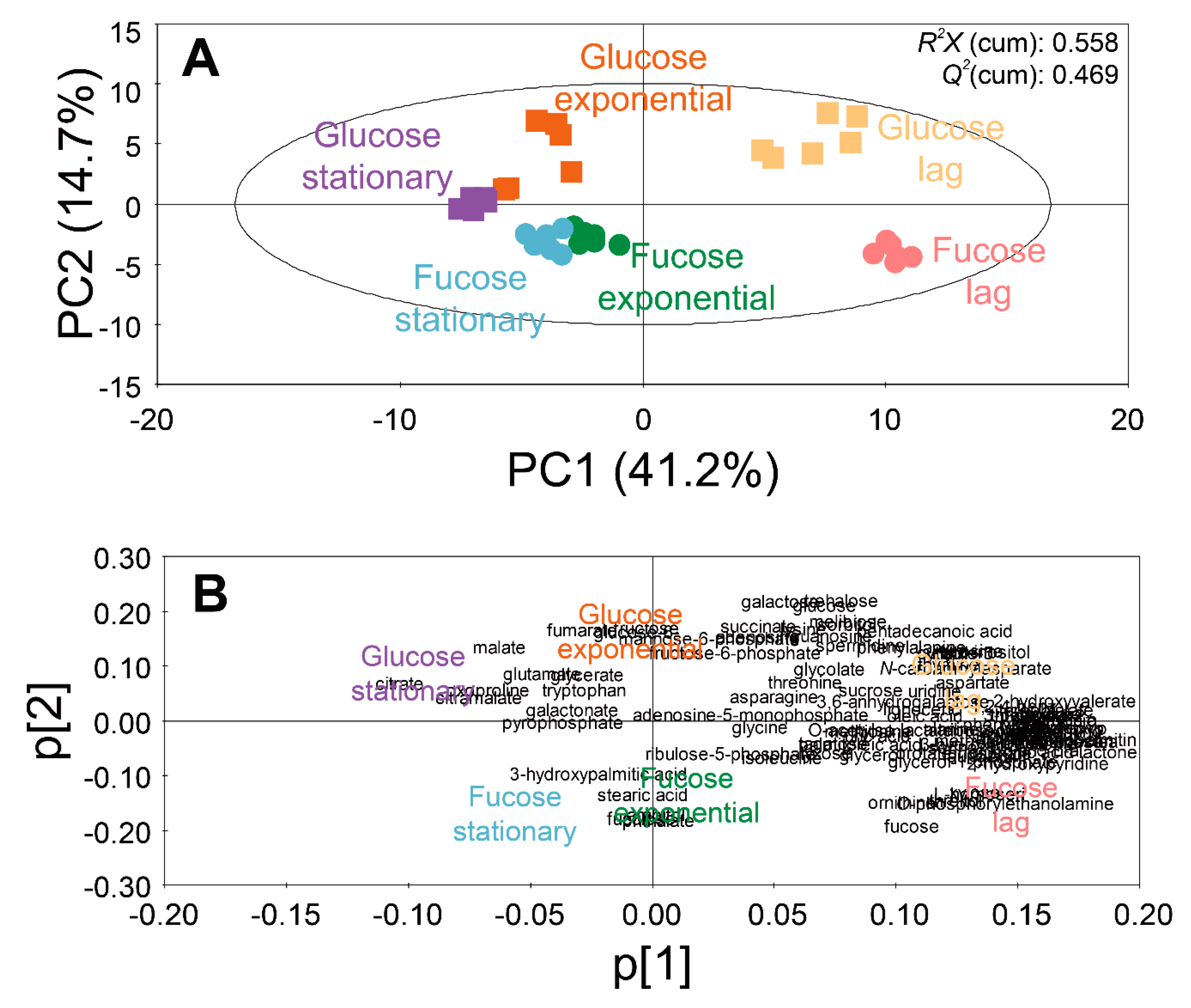
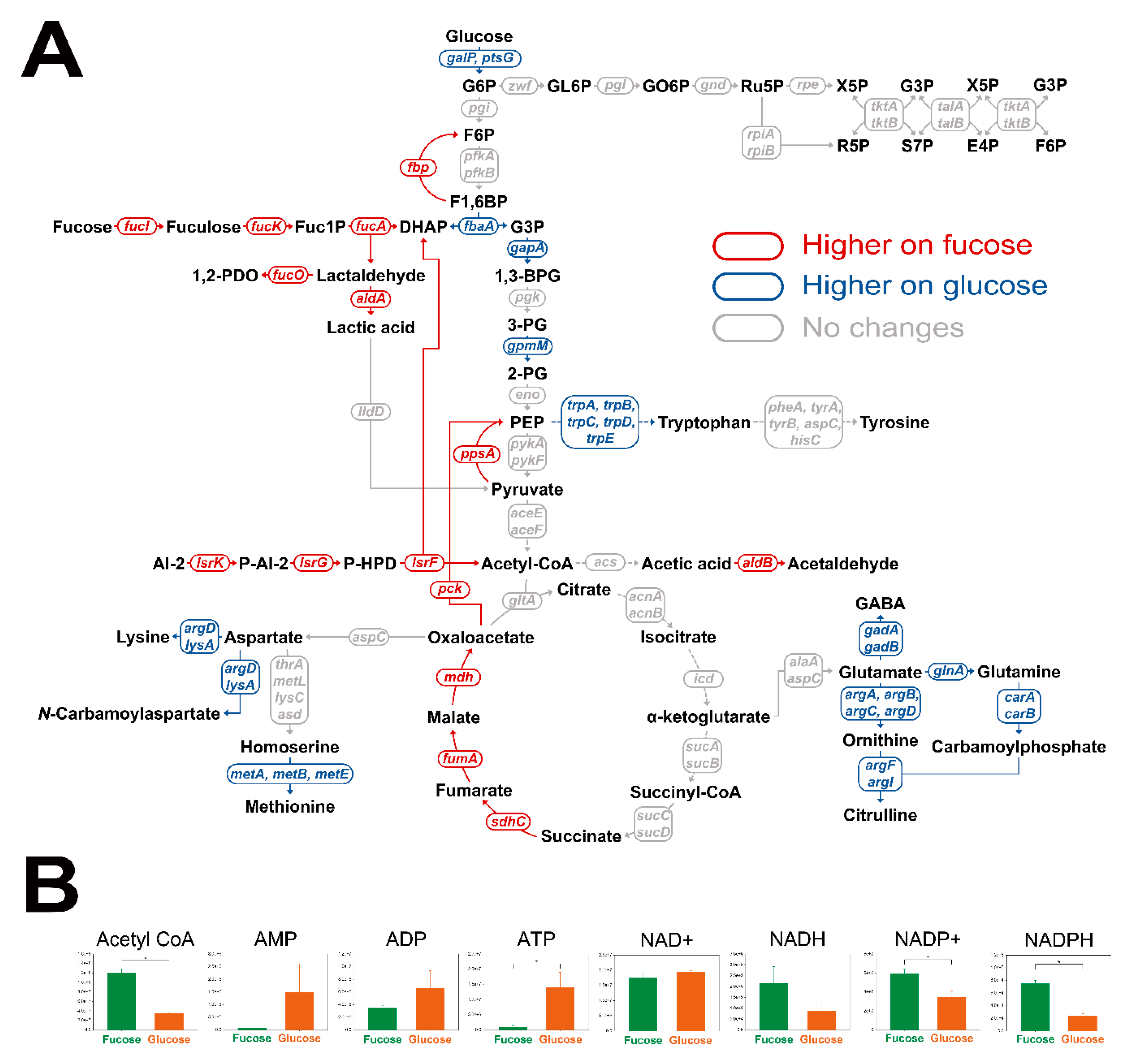
2.3. Comparison of Intracellular Metabolite Profiles of E. coli on Fucose and Glucose
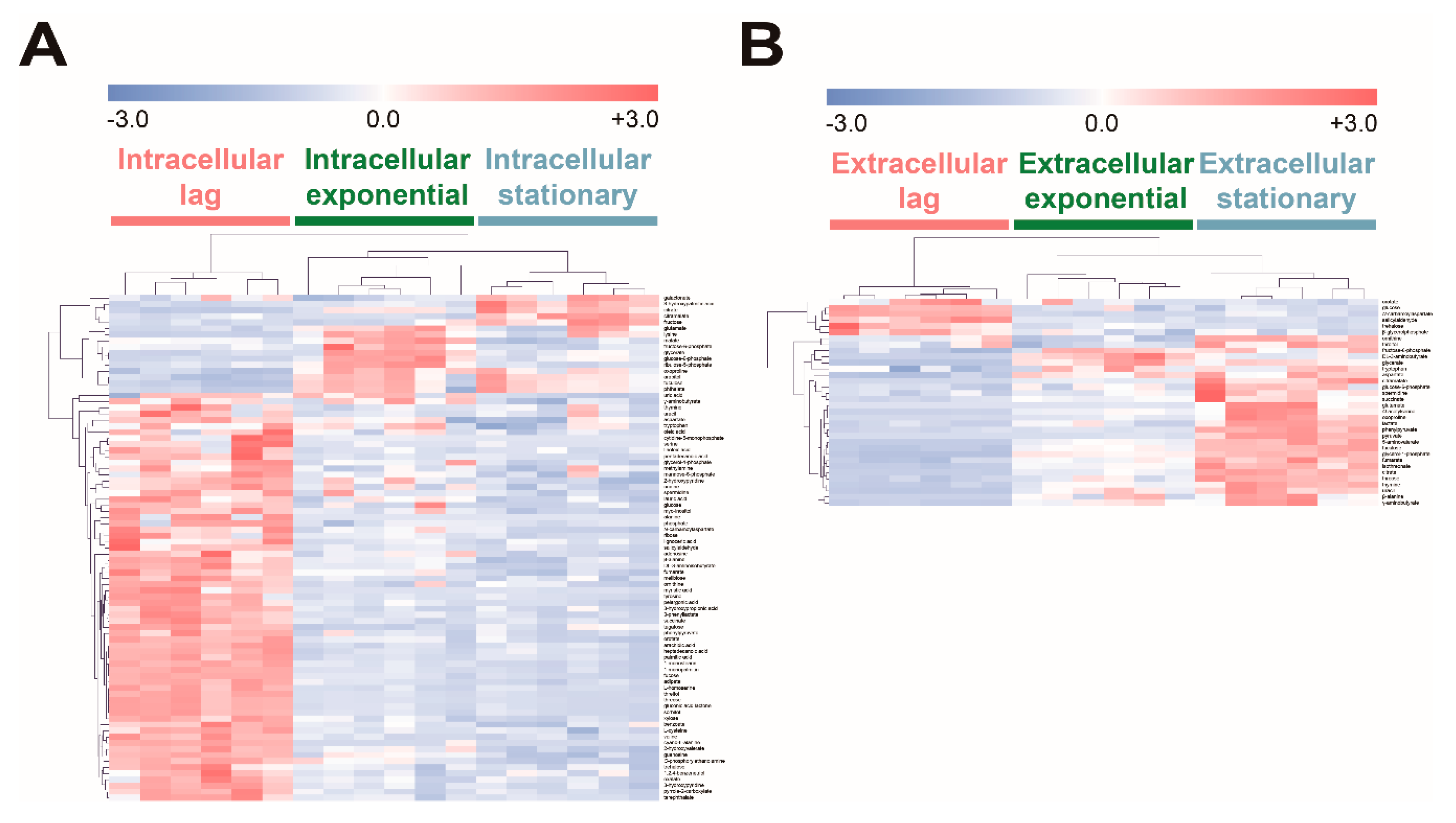
2.4. Comparison of Intracellular and Extracellular Metabolite Profiles of E. coli on Fucose at Different Growth Phases
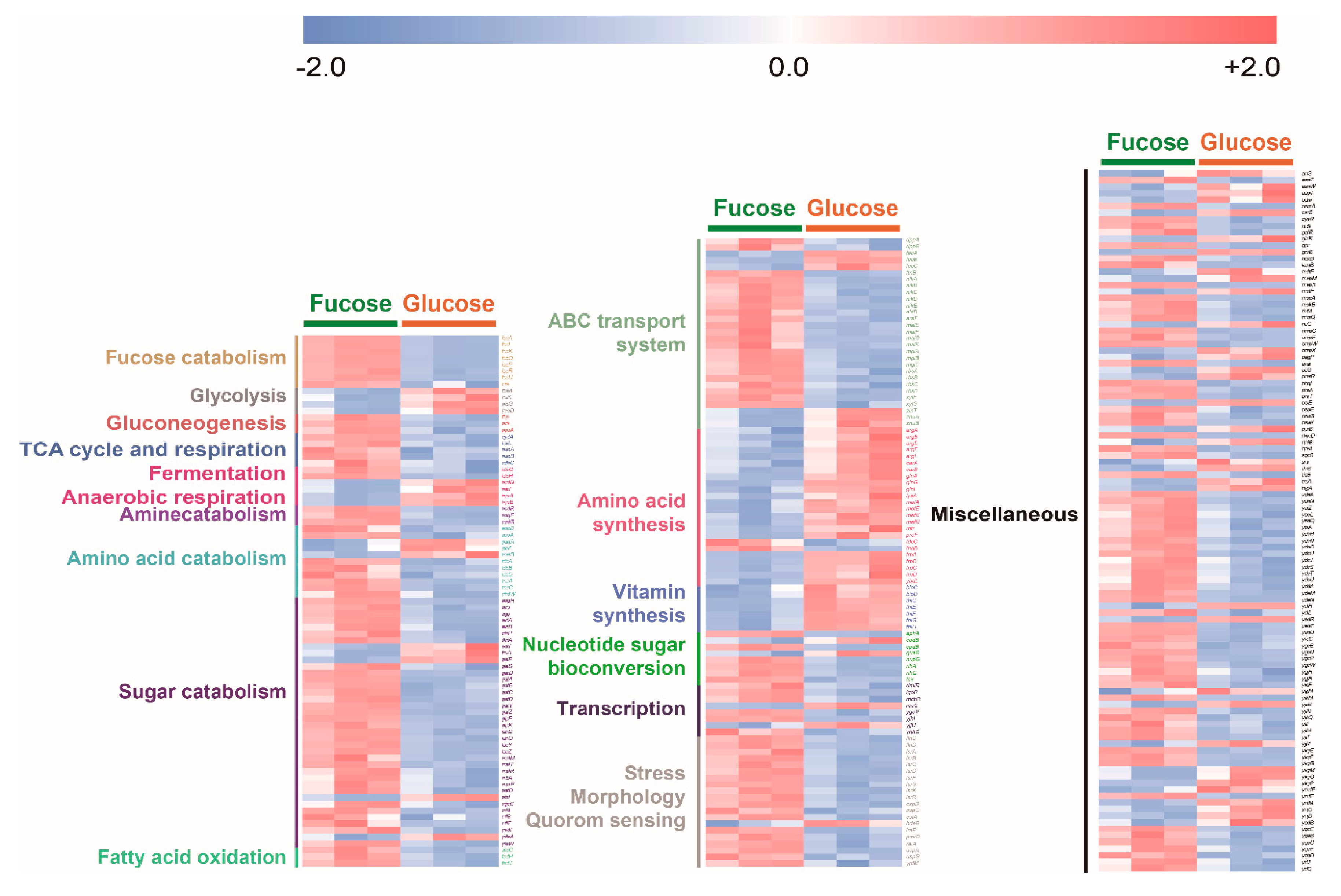
2.5. Comparison of Transcriptome and Cofactor Profiles of E. coli on Fucose and Glucose in the Exponential Phase
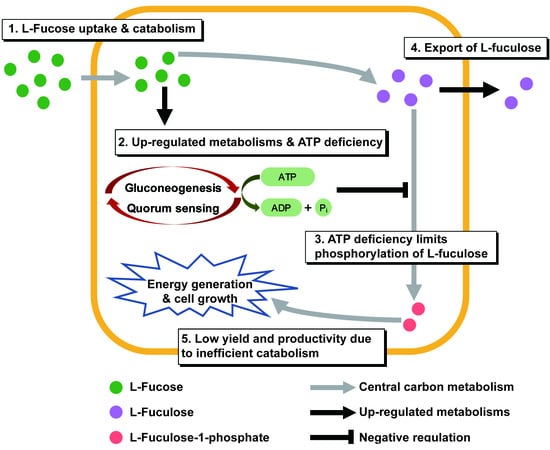
3. Discussion
4. Materials and Methods
4.1. Strains, Media, and Culture Conditions
4.2. Preparation of Intracellular and Extracellular Metabolite Samples
4.3. GC/TOF–MS and HPLC Analyses of Intracellular and Extracellular Metabolites
4.4. GC/TOF–MS Data Processing and Statistical Analyses
4.5. Quality Controls for GC/TOF–MS Analysis
4.6. LC/MS Analysis of Energy and CoA Metabolites
4.7. RNA Isolation and RNA Library Construction
4.8. Statistical Analysis of Gene Expression Levels
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Keasling, J.D.; Chou, H. Metabolic engineering delivers next-generation biofuels. Nat. Biotechnol. 2008, 26, 298–299. [Google Scholar] [CrossRef] [PubMed]
- Alonso, D.M.; Bond, J.Q.; Dumesic, J.A. Catalytic conversion of biomass to biofuels. Green Chem. 2010, 12, 1493–1513. [Google Scholar] [CrossRef]
- Peralta-Yahya, P.P.; Zhang, F.; del Cardayre, S.B.; Keasling, J.D. Microbial engineering for the production of advanced biofuels. Nature 2012, 488, 320–328. [Google Scholar] [CrossRef]
- Wei, N.; Quarterman, J.; Jin, Y.S. Marine macroalgae: An untapped resource for producing fuels and chemicals. Trends Biotechnol. 2013, 31, 70–77. [Google Scholar] [CrossRef]
- Andriamanantoanina, H.; Rinaudo, M. Characterization of the alginates from five madagascan brown algae. Carbohyd. Polym. 2010, 82, 555–560. [Google Scholar] [CrossRef]
- Wu, S.; Sun, J.; Chi, S.; Wang, L.; Wang, X.; Liu, C.; Li, X.; Yin, J.; Liu, T.; Yu, J. Transcriptome sequencing of essential marine brown and red algal species in China and its significance in algal biology and phylogeny. Acta Oceanol. Sin. 2014, 33, 1–12. [Google Scholar] [CrossRef]
- Roesijadi, G.; Jones, S.B.; Snowden-Swan, L.J.; Zhu, Y. Macroalgae as a Biomass Feedstock: A Preliminary Analysis; Pacific Northwest National Lab. (PNNL): Richland, WA, USA, 2010.
- Wang, D.; Kim, D.H.; Kim, K.H. Effective production of fermentable sugars from brown macroalgae biomass. Appl. Microbiol. Biotechnol. 2016, 100, 9439–9450. [Google Scholar] [CrossRef]
- Wijesinghe, W.; Jeon, Y.J. Biological activities and potential industrial applications of fucose rich sulfated polysaccharides and fucoidans isolated from brown seaweeds: A review. Carbohyd. Polym. 2012, 88, 13–20. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, D.; Luo, G.; Zhang, S.; Chen, J. Macroalgae for biofuels production: Progress and perspectives. Renew. Sust. Energ. Rev. 2015, 47, 427–437. [Google Scholar] [CrossRef]
- Saxena, R.K.; Anand, P.; Saran, S.; Isar, J.; Agarwal, L. Microbial production and applications of 1,2-propanediol. Indian J. Microbiol. 2010, 50, 2–11. [Google Scholar] [CrossRef]
- Danese, P.N.; Pratt, L.A.; Kolter, R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 2000, 182, 3593–3596. [Google Scholar] [CrossRef] [PubMed]
- Han, N.S.; Kim, T.J.; Park, Y.C.; Kim, J.; Seo, J.H. Biotechnological production of human milk oligosaccharides. Biotechnol. Adv. 2012, 30, 1268–1278. [Google Scholar] [CrossRef]
- Chin, Y.W.; Kim, J.Y.; Lee, W.H.; Seo, J.H. Enhanced production of 2′-fucosyllactose in engineered Escherichia coli BL21star(DE3) by modulation of lactose metabolism and fucosyltransferase. J. Biotechnol. 2015, 210, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Yang, K.; Liu, J.; Xu, Y.; Wang, Y.; Wang, R.; Liu, B.; Feng, L. 3-fucosyllactose through modular pathway enhancement. Metab. Eng. 2017, 41, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Nielsen, J. Advancing metabolic engineering through systems biology of industrial microorganisms. Curr. Opin. Biotechnol. 2015, 36, 8–15. [Google Scholar] [CrossRef]
- Chiu, T.H.; Feingold, D.S. L-rhamnulose 1-phosphate aldolase from Escherichia coli. Crystallization and properties. Biochemistry 1969, 8, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Hacking, A.J.; Lin, E.C. Disruption of the fucose pathway as a consequence of genetic adaptation to propanediol as a carbon source in Escherichia coli. J. Bacteriol. 1976, 126, 1166–1172. [Google Scholar] [PubMed]
- Baldoma, L.; Aguilar, J. Metabolism of L-fucose and L-rhamnose in Escherichia coli: Aerobic-anaerobic regulation of L-lactaldehyde dissimilation. J. Bacteriol. 1988, 170, 416–421. [Google Scholar] [CrossRef]
- Becerra, J.E.; Yebra, M.J.; Monedero, V. An L-fucose operon in the probiotic Lactobacillus rhamnosus GG is involved in adaptation to gastrointestinal conditions. Appl. Environ. Microbiol. 2015, 81, 3880–3888. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.; Stark, H.; Fafenrot, K.; Albersmeier, A.; Pham, T.K.; Muller, K.B.; Meyer, B.H.; Hoffmann, L.; Shen, L.; Albaum, S.P.; et al. A systems biology approach reveals major metabolic changes in the thermoacidophilic archaeon Sulfolobus solfataricus in response to the carbon source L-fucose versus D-glucose. Mol. Microbiol. 2016, 102, 882–908. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, L.; Tian, K.; Kumar, A.; Singh, S.; Prior, B.A.; Wang, Z. Metabolic engineering of Escherichia coli: A sustainable industrial platform for bio-based chemical production. Biotechnol. Adv. 2013, 31, 1200–1223. [Google Scholar] [CrossRef]
- Rochfort, S. Metabolomics reviewed: A new “Omics” platform technology for systems biology and implications for natural products research. J. Nat. Prod. 2005, 68, 1813–1820. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, F.; Nie, L. Integrating multiple ’omics’ analysis for microbial biology: Application and methodologies. Microbiol-Sgm 2010, 156, 287–301. [Google Scholar] [CrossRef]
- Kim, M.; Rai, N.; Zorraquino, V.; Tagkopoulos, I. Multi-omics integration accurately predicts cellular state in unexplored conditions for Escherichia coli. Nat. Commun. 2016, 7, 13090. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Scalbert, A.; Brennan, L.; Fiehn, O.; Hankemeier, T.; Kristal, B.S.; Ommen, B.V.; Pujos-Guillot, E.; Verheij, E.; Wishart, D.; Wopereis, S. Mass-spectrometry-based metabolomics: Limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics 2009, 5, 435–458. [Google Scholar] [CrossRef]
- Gonzalez, R.; Tao, H.; Shanmugam, K.T.; York, S.W.; Ingram, L.O. Global gene expression differences associated with changes in glycolytic flux and growth rate in Escherichia coli during the fermentation of glucose and xylose. Biotechnol. Progr. 2002, 18, 6–20. [Google Scholar] [CrossRef]
- Shimizu, K. Metabolic regulation and coordination of the metabolism in bacteria in response to a variety of growth conditions. In Advances in Biochemical Engineering/Biotechnology; Ye, Q., Bao, J., Zhong, J.J., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 1–54. [Google Scholar]
- Scott, K.P.; Martin, J.C.; Campbell, G.; Mayer, C.D.; Flint, H.J. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans”. J. Bacteriol. 2006, 188, 4340–4349. [Google Scholar] [CrossRef]
- Fujiya, M.; Musch, M.W.; Nakagawa, Y.; Hu, S.; Alverdy, J.; Kohgo, Y.; Schneewind, O.; Jabri, B.; Chang, E.B. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe 2007, 1, 299–308. [Google Scholar] [CrossRef]
- Marques, J.C.; Oh, I.K.; Ly, D.C.; Lamosa, P.; Ventura, M.R.; Miller, S.T.; Xavier, K.B. LsrF, a coenzyme A-dependent thiolase, catalyzes the terminal step in processing the quorum sensing signal autoinducer-2. Proc. Natl. Acad. Sci. USA 2014, 111, 14235–14240. [Google Scholar] [CrossRef]
- De Mey, M.; De Maeseneire, S.; Soetaert, W.; Vandamme, E. Minimizing acetate formation in E. coli fermentations. J. Ind. Microbiol. Biotechnol. 2007, 34, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Quan, C.; Jin, L.; Chen, M. Production, detection and application perspectives of quorum sensing autoinducer-2 in bacteria. J. Biotechnol. 2018, 268, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, D.Y.; Wohlgemuth, G.; Park, H.S.; Fiehn, O.; Kim, K.H. Evaluation and optimization of metabolome sample preparation methods for Saccharomyces cerevisiae. Anal. Chem. 2013, 85, 2169–2176. [Google Scholar] [CrossRef] [PubMed]
- Skogerson, K.; Wohlgemuth, G.; Barupal, D.K.; Fiehn, O. The volatile compound BinBase mass spectral database. BMC Bioinform. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef]
- Barupal, D.K.; Haldiya, P.K.; Wohlgemuth, G.; Kind, T.; Kothari, S.L.; Pinkerton, K.E.; Fiehn, O. MetaMapp: Mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinform. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Bhagabati, N.K.; Braisted, J.C.; Liang, W.; Sharov, V.; Howe, E.A.; Li, J.; Thiagarajan, M.; White, J.A.; Quackenbush, J. TM4 microarray software suite. Methods Enzymol. 2006, 411, 134–193. [Google Scholar]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Lee, D.Y.; Fiehn, O. High quality metabolomic data for Chlamydomonas reinhardtii. Plant Methods 2008, 4. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Cheong, Y.E.; Jung, I.; Kim, K.H. Metabolomic and Transcriptomic Analyses of Escherichia coli for Efficient Fermentation of L-Fucose. Mar. Drugs 2019, 17, 82. https://doi.org/10.3390/md17020082
Kim J, Cheong YE, Jung I, Kim KH. Metabolomic and Transcriptomic Analyses of Escherichia coli for Efficient Fermentation of L-Fucose. Marine Drugs. 2019; 17(2):82. https://doi.org/10.3390/md17020082
Chicago/Turabian StyleKim, Jungyeon, Yu Eun Cheong, Inho Jung, and Kyoung Heon Kim. 2019. "Metabolomic and Transcriptomic Analyses of Escherichia coli for Efficient Fermentation of L-Fucose" Marine Drugs 17, no. 2: 82. https://doi.org/10.3390/md17020082
APA StyleKim, J., Cheong, Y. E., Jung, I., & Kim, K. H. (2019). Metabolomic and Transcriptomic Analyses of Escherichia coli for Efficient Fermentation of L-Fucose. Marine Drugs, 17(2), 82. https://doi.org/10.3390/md17020082