Saline Environments as a Source of Potential Quorum Sensing Disruptors to Control Bacterial Infections: A Review
Abstract
:1. Introduction
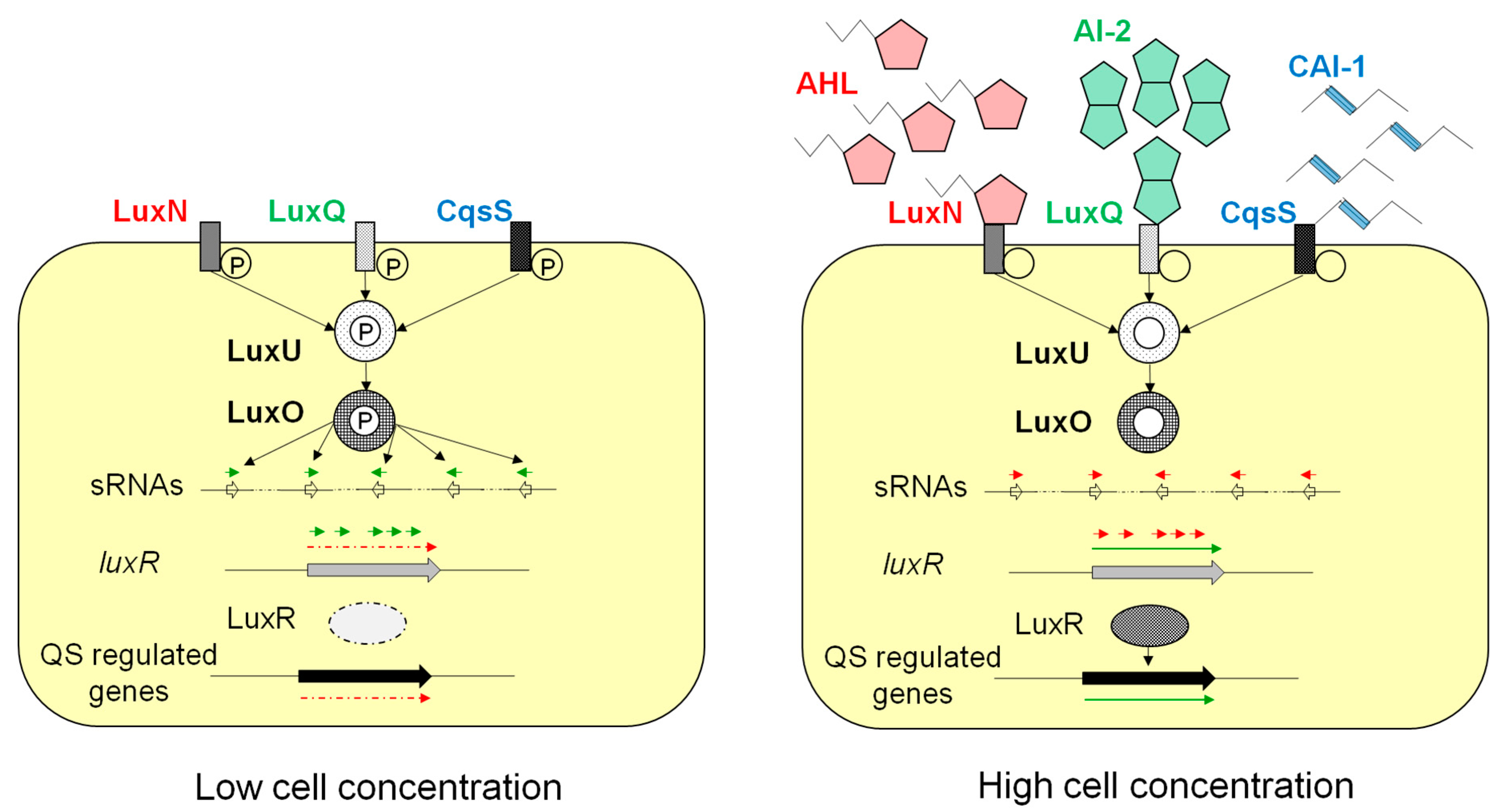
2. Quorum Sensing in Bacteria of Aquacultural Importance
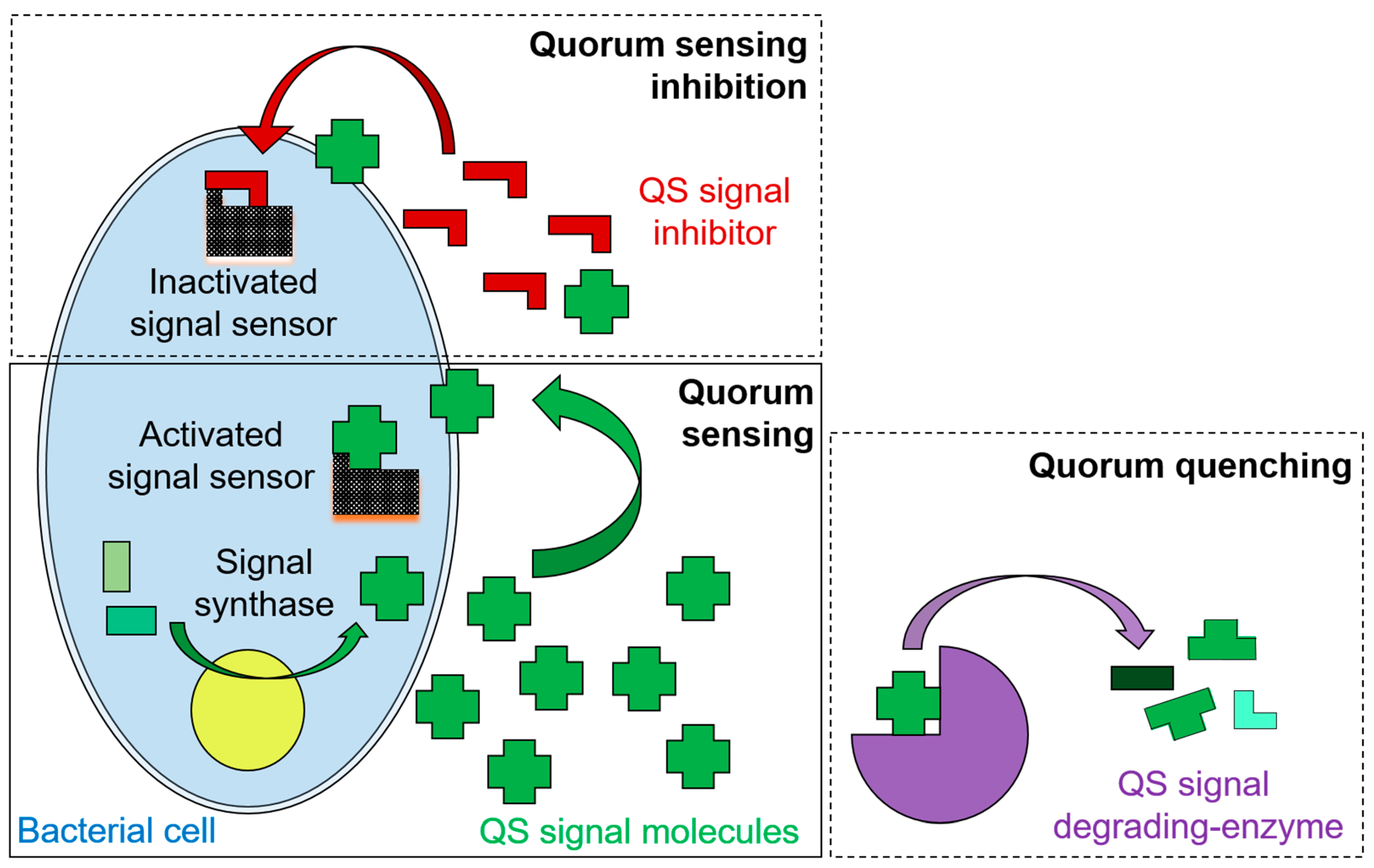
3. Inhibition of Quorum Sensing Mechanisms
4. Saline Environments as an Important Source of Bioactive Molecules
5. Quorum Sensing Interference in Marine Environments
6. Quorum Sensing Interference in Saline and Hypersaline Environments
7. Applications in Aquaculture and Other Industries
Conflicts of Interest
References
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial quorum sensing and microbial community interactions. MBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Urbanczyk, H.; Ast, J.C.; Higgins, M.J.; Carson, J.; Dunlap, P.V. Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 2823–2829. [Google Scholar] [CrossRef]
- Eberhard, A. Inhibition and activation of bacterial luciferase synthesis. J. Bacteriol. 1972, 109, 1101–1105. [Google Scholar]
- Eberhard, A.; Burlingame, A.L.; Eberhard, C.; Kenyon, G.L.; Nealson, K.H.; Oppenheimer, N.J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 1981, 20, 2444–2449. [Google Scholar] [CrossRef]
- Zarubin, M.; Belkin, S.; Ionescu, M.; Genin, A. Bacterial bioluminescence as a lure for marine zooplankton and fish. Proc. Natl. Acad. Sci. USA 2012, 109, 853–857. [Google Scholar] [CrossRef]
- Dunn, A.K. Vibrio fischeri metabolism. Adv. Microb. Physiol. 2012, 61, 37–68. [Google Scholar] [CrossRef]
- Ng, W.W.L.; Bassler, B.B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef]
- Parker, C.T.; Sperandio, V. Cell-to-cell signalling during pathogenesis. Cell. Microbiol. 2009, 11, 363–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, G.; Ray, A.K. The talking language in some major Gram-negative bacteria. Arch. Microbiol. 2016, 198, 489–499. [Google Scholar] [CrossRef]
- Flavier, A.B.; Clough, S.J.; Schell, M.A.; Denny, T.P. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 1997, 26, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Holden, M.T.G.; Chhabra, S.R.; De Nys, R.; Stead, P.; Bainton, N.J.; Hill, P.J.; Manefield, M.; Kumar, N.; Labatte, M.; England, D.; et al. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol. Microbiol. 1999, 33, 1254–1266. [Google Scholar] [CrossRef]
- Deziel, E.; Lepine, F.; Milot, S.; He, J.; Mindrinos, M.N.; Tompkins, R.G.; Rahme, L.G. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA 2004, 101, 1339–1344. [Google Scholar] [CrossRef]
- Barber, C.E.; Tang, J.L.; Feng, J.X.; Pan, M.Q.; Wilson, T.J.; Slater, H.; Dow, J.M.; Williams, P.; Daniels, M.J. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 1997, 24, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Brameyer, S.; Kresovic, D.; Bode, H.B.; Heermann, R. Dialkylresorcinols as bacterial signaling molecules. Proc. Natl. Acad. Sci. USA 2015, 112, 572–577. [Google Scholar] [CrossRef]
- Gospodarek, E.; Bogiel, T.; Zalas-Wiecek, P. Communication between microorganisms as a basis for production of virulence factors. Polish J. Microbiol. 2009, 58, 191–198. [Google Scholar]
- Dessaux, Y.; Faure, D. Quorum sensing and quorum quenching in Agrobacterium: A “go/no go system”? Genes (Basel) 2018, 9, 210. [Google Scholar] [CrossRef]
- García-Aljaro, C.; Vargas-Cespedes, G.J.; Blanch, A.R. Detection of acylated homoserine lactones produced by Vibrio spp. and related species isolated from water and aquatic organisms. J. Appl. Microbiol. 2011, 112, 383–389. [Google Scholar] [CrossRef]
- Rasmussen, B.B.; Nielsen, K.F.; MacHado, H.; Melchiorsen, J.; Gram, L.; Sonnenschein, E.C. Global and phylogenetic distribution of quorum sensing signals, acyl homoserine lactones, in the family of Vibrionaceae. Mar. Drugs 2014, 12, 5527–5546. [Google Scholar] [CrossRef]
- Yang, Q.; Han, Y.; Zhang, X.H. Detection of quorum sensing signal molecules in the family Vibrionaceae. J. Appl. Microbiol. 2011, 110, 1438–1448. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kuang, W.; Long, L.; Zhang, S. Production of quorum-sensing signals by bacteria in the coral mucus layer. Coral Reefs 2017, 36, 1235–1241. [Google Scholar] [CrossRef]
- Liu, J.; Fu, K.; Wu, C.; Qin, K.; Li, F.; Zhou, L. “In-group” communication in marine Vibrio: A review of n-acyl homoserine lactones-driven quorum sensing. Front. Cell. Infect. Microbiol. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.R.; Liu, P.C.; Lee, K.K. Lethal attribute of serine protease secreted by Vibrio alginolyticus strains in Kuruma prawn Penaeus japonicus. Z. Naturforsch. C 2000, 55, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Prado, S.; Dubert, J.; Barja, J.L. Characterization of pathogenic vibrios isolated from bivalve hatcheries in Galicia, NW Atlantic coast of Spain. Description of Vibrio tubiashii subsp. europaensis subsp. nov. Syst. Appl. Microbiol. 2015, 38, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Gómez-León, J.; Villamil, L.; Lemos, M.L.; Novoa, B.; Figueras, A. Isolation of Vibrio alginolyticus and Vibrio splendidus from aquacultured carpet shell clam (Ruditapes decussatus) larvae associated with mass mortalities. Appl. Environ. Microbiol. 2005, 71, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Rojas, R.; Miranda, C.D.; Santander, J.; Romero, J. First report of Vibrio tubiashii associated with a massive larval mortality event in a commercial hatchery of scallop Argopecten purpuratus in Chile. Front. Microbiol. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- Dubert, J.; Barja, J.L.; Romalde, J.L. New insights into pathogenic vibrios affecting bivalves in hatcheries: present and future prospects. Front. Microbiol. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Ruwandeepika, H.A.D.; Defoirdt, T.; Bhowmick, P.P.; Karunasagar, I.; Karunasagar, I.; Bossier, P. In vitro and in vivo expression of virulence genes in Vibrio isolates belonging to the Harveyi clade in relation to their virulence towards gnotobiotic brine shrimp (Artemia franciscana). Environ. Microbiol. 2011, 13, 506–517. [Google Scholar] [CrossRef]
- Bassler, B.B.L.; Wright, M.; Silverman, M.M.R. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: Sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 1994, 13, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.L.; Hughson, F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002, 415, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Skorupski, K.; Lenz, D.H.; Taylor, R.K.; Bassler, B.L. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 2002, 110, 303–314. [Google Scholar] [CrossRef]
- Higgins, D.A.; Pomianek, M.E.; Kraml, C.M.; Taylor, R.K.; Semmelhack, M.F.; Bassler, B.L. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 2007, 450, 883–886. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.L.; Perez, L.J.; Wei, Y.; Kraml, C.; Semmelhack, M.F.; Bassler, B.L. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol. Microbiol. 2011, 79, 1407–1417. [Google Scholar] [CrossRef]
- Milton, D.L. Quorum sensing in vibrios: Complexity for diversification. Int. J. Med. Microbiol. 2006, 296, 61–71. [Google Scholar] [CrossRef]
- Defoirdt, T. Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 2018, 26, 313–328. [Google Scholar] [CrossRef]
- Croxatto, A.; Pride, J.; Hardman, A.; Williams, P.; Cámara, M.; Milton, D. A distinctive dual-channel quorum sensing system operates in Vibrio anguillarum. Mol. Microbiol. 2004, 52, 1677–1689. [Google Scholar] [CrossRef]
- Morohoshi, T.; Inaba, T.; Kato, N.; Kanai, K.; Ikeda, T. Identification of quorum-sensing signal molecules and the LuxRI homologs in fish pathogen Edwardsiella tarda. J. Biosci. Bioeng. 2004, 98, 274–281. [Google Scholar] [CrossRef]
- Bruhn, J.B.; Dalsgaard, I.; Nielsen, K.F.; Buchholtz, C.; Larsen, J.L.; Gram, L. Quorum sensing signal molecules (acylated homoserine lactones) in Gram-negative fish pathogenic bacteria. Dis. Aquat. Organ. 2005, 65, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Defoirdt, T.; Bossier, P.; Sorgeloos, P.; Verstraete, W. The impact of mutations in the quorum sensing systems of Aeromonas hydrophila, Vibrio anguillarum and Vibrio harveyi on their virulence towards gnotobiotically cultured Artemia franciscana. Environ. Microbiol. 2005, 7, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fu, K.; Wang, Y.; Wu, C.; Li, F.; Shi, L.; Ge, Y.; Zhou, L. Detection of diverse N-acyl-homoserine lactones in Vibrio alginolyticus and regulation of biofilm formation by N-(3-oxodecanoyl) homoserine lactone in vitro. Front. Microbiol. 2017, 8, 1–15. [Google Scholar] [CrossRef]
- Li, X.; Dierckens, K.; Bossier, P.; Defoirdt, T. The impact of quorum sensing on the virulence of Vibrio anguillarum towards gnotobiotic sea bass (Dicentrarchus labrax) larvae. Aquac. Res. 2018, 49, 3686–3689. [Google Scholar] [CrossRef]
- Defoirdt, T.; Verstraete, W.; Bossier, P. Luminescence, virulence and quorum sensing signal production by pathogenic Vibrio campbellii and Vibrio harveyi isolates. J. Appl. Microbiol. 2008, 104, 1480–1487. [Google Scholar] [CrossRef] [PubMed]
- Lilley, B.N.; Bassler, B.L. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 2000, 36, 940–954. [Google Scholar] [CrossRef]
- Milton, D.L.; Hardman, A.; Camara, M.; Chhabra, S.R.; Bycroft, B.W.; Stewart, G.S.A.B.; Williams, P. Quorum sensing in Vibrio anguillarum: characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-L-homoserine lactone. J. Bacteriol. 1997, 179, 3004–3012. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Reina, J.C.; Fuentes-Monteverde, J.C.; Fernandez, G.; Rodrıguez, J.; Llamas, I. AHL-lactonase expression in three marine emerging pathogenic Vibrio spp. reduces virulence and mortality in brine shrimp (Artemia salina) and Manila clam (Venerupis philippinarum). PLoS ONE 2018, 13, e0195176. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T.; Boon, N.; Sorgeloos, P.; Verstraete, W.; Bossier, P. Quorum sensing and quorum quenching in Vibrio harveyi: Lessons learned from in vivo work. ISME J. 2008, 2, 19–26. [Google Scholar] [CrossRef]
- Finch, R. Quorum sensing: A novel target for anti-infective therapy. J. Antimicrob. Chemother. 1998, 42, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.H.H. Quorum quenching and proactive host defense. Trends Plant Sci. 2003, 8, 238–244. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, L. Quorum sensing and quorum-quenching enzymes. J. Microbiol. 2005, 43, 101–109. [Google Scholar] [PubMed]
- Czajkowski, R.; Jafra, S. Quenching of acyl-homoserine lactone-dependent quorum sensing by enzymatic disruption of signal molecules. Acta Biochim. Pol. 2009, 56, 1–16. [Google Scholar] [CrossRef]
- Uroz, S.; Dessaux, Y.; Oger, P. Quorum sensing and quorum quenching: The yin and yang of bacterial communication. ChemBioChem 2009, 10, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.W.; Koh, C.L.; Sam, C.K.; Yin, W.F.; Chan, K.G. Quorum quenching revisited: From signal decays to signalling confusion. Sensors 2012, 12, 4661–4696. [Google Scholar] [CrossRef] [PubMed]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum quenching: role in nature and applied developments. FEMS Microbiol. Rev. 2016, 40, 86–116. [Google Scholar] [CrossRef] [PubMed]
- Cegelski, L.; Marshall, G.R.; Eldridge, G.R.; Hultgren, S.J. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 2008, 6, 17–27. [Google Scholar] [CrossRef]
- Zucca, M.; Scutera, S.; Savoia, D. New antimicrobial frontiers. Mini Rev. Med. Chem. 2011, 11, 888–900. [Google Scholar] [CrossRef]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef]
- Defoirdt, T.; Boon, N.; Bossier, P. Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 2010, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- García-Contreras, R.; Maeda, T.; Wood, T.K. Resistance to quorum-quenching compounds. Appl. Environ. Microbiol. 2013, 79, 6840–6846. [Google Scholar] [CrossRef]
- García-Contreras, R.; Maeda, T.; Wood, T.K. Can resistance against quorum-sensing interference be selected? ISME J. 2016, 1–7. [Google Scholar] [CrossRef]
- Kalia, V.C. Quorum sensing inhibitors: An overview. Biotechnol. Adv. 2013, 31, 224–245. [Google Scholar] [CrossRef]
- Kalia, V.C.; Wood, T.K.; Kumar, P. Evolution of resistance to quorum-sensing inhibitors. Microb. Ecol. 2014, 68, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Sandoz, K.M.; Mitzimberg, S.M.; Schuster, M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. USA 2007, 104, 15876–15881. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; García-Contreras, R.; Pu, M.; Sheng, L.; Garcia, L.R.; Tomás, M.; Wood, T.K. Quorum quenching quandary: resistance to antivirulence compounds. ISME J. 2012, 6, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Mellbye, B.; Schuster, M. The sociomicrobiology of antivirulence drug resistance: A proof of concept. MBio 2011, 2, e00131-11. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T.; Sorgeloos, P.; Bossier, P. Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr. Opin. Microbiol. 2011, 14, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Rémy, B.; Plener, L.; Elias, M.; Daudé, D.; Chabrière, E. Des enzymes pour bloquer la communication bactérienne, une alternative aux antibiotiques? Ann. Pharm. Françaises 2016, 74, 413–420. [Google Scholar] [CrossRef]
- Bzdrenga, J.; Daudé, D.; Rémy, B.; Jacquet, P.; Plener, L.; Elias, M.; Chabrière, E. Biotechnological applications of quorum quenching enzymes. Chem. Biol. Interact. 2017, 1, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.H.H.; Xu, J.L.; Li, X.Z.; Zhang, L.H. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 2000, 97, 3526–3531. [Google Scholar] [CrossRef]
- Dong, Y.H.; Wang, L.H.; Xu, J.L.; Zhang, H.B.; Zhang, X.F.; Zhang, L.H. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 2001, 411, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Gusti, A.; Zhang, Q.; Xu, J.; Zhang, L. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl. Environ. Microbiol. 2002, 68, 1754–1759. [Google Scholar] [CrossRef] [PubMed]
- Leadbetter, J.R.; Greenberg, E.P. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 2000, 182, 6921–6926. [Google Scholar] [CrossRef]
- Uroz, S.; D’Angelo-Picard, C.; Carlier, A.; Elasri, M.; Sicot, C.; Petit, A.; Oger, P.; Faure, D.; Dessaux, Y. Novel bacteria degrading N-acylhomoserine lactones and their use as quenchers of quorum-sensing-regulated functions of plant-pathogenic bacteria. Microbiology 2003, 149, 1981–1989. [Google Scholar] [CrossRef]
- Hornby, J.M.; Jensen, E.C.; Lisec, A.D.; Tasto, J.J.; Jahnke, B.; Shoemaker, R.; Dussault, P.; Nickerson, K.W. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 2001, 67, 2982–2992. [Google Scholar] [CrossRef]
- Wong, C.S.; Koh, C.L.; Sam, C.K.; Chen, J.; Chong, Y.; Yin, W.F.; Chan, K.G. Degradation of bacterial quorum sensing signaling molecules by the microscopic yeast Trichosporon loubieri isolated from tropical wetland waters. Sensors 2013, 13, 12943–12957. [Google Scholar] [CrossRef]
- Leguina, A.C.d.V.; Nieto, C.; Pajot, H.F.; Bertini, E.V.; Mac Cormack, W.; Castellanos de Figueroa, L.I.; Nieto-Peñalver, C.G. Inactivation of bacterial quorum sensing signals N-acyl homoserine lactones is widespread in yeasts. Fungal Biol. 2018, 122, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.B.; Skindersoe, M.E.; Bjarnsholt, T.; Phipps, R.K.; Christensen, K.B.; Jensen, P.O.; Andersen, J.B.; Koch, B.; Larsen, T.O.; Hentzer, M.; et al. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 2016, 151, 1325–1340. [Google Scholar] [CrossRef]
- Uroz, S.; Heinonsalo, J. Degradation of N-acyl homoserine lactone quorum sensing signal molecules by forest root-associated fungi. FEMS Microbiol. Ecol. 2008, 65, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Givskov, M.; de Nys, R.; Manefield, M.; Gram, L.; Maximilien, R.; Eberl, L.; Molin, S.; Steinberg, P.D.; Kjelleberg, S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bact. 1996, 178, 6618–6622. [Google Scholar] [CrossRef]
- Teplitski, M.; Robinson, J.B.; Bauer, W.D. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant-Microbe Interact. 2000, 13, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Huber, B.; Eberl, L.; Feucht, W.; Polster, J. Influence of polyphenols on bacterial biofilm formation and quorum sensing. Z. Naturforsch. C 2003, 58, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Delalande, L.; Faure, D.; Raffoux, A.A.; Uroz, S.S.; D’Angelo-Picard, C.; Elasri, M.; Carlier, A.A.; Berruyer, R.; Petit, A.; Williams, P.; et al. N-hexanoyl-l-homoserine lactone, a mediator of bacterial quorum-sensing regulation, exhibits plant-dependent stability and may be inactivated by germinating Lotus corniculatus seedlings. FEMS Microbiol. Ecol. 2005, 52, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Draganov, D.I.; Teiber, J.F.; Speelman, A.; Osawa, Y.; Sunahara, R.; La Du, B.N. Human paraoxonases (PON1, PON2 and PON3) are lactonases with overlapping and distinct substrate specificities. J. Lipid Res. 2005, 46, 1239–1247. [Google Scholar] [CrossRef]
- Yang, F.; Wang, L.H.; Wang, J.; Dong, Y.H.; Hu, J.Y.; Zhang, L.H. Quorum quenching enzyme activity is widely conserved in the sera of mammalian species. FEBS Lett. 2005, 579, 3713–3717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Kaufmann, G.F.; Bowen, J.P.; Arbiser, J.L.; Janda, K.D. Solenopsin A, a venom alkaloid from the fire ant Solenopsis invicta, inhibits quorum-sensing signaling in Pseudomonas aeruginosa. J. Infect. Dis. 2008, 198, 1198–1201. [Google Scholar] [CrossRef]
- Kalia, V.C.; Purohit, H.J. Quenching the quorum sensing system: Potential antibacterial drug targets. Crit. Rev. Microbiol. 2011, 37, 121–140. [Google Scholar] [CrossRef]
- Bhardwaj, A.K.; Vinothkumar, K.; Rajpara, N. Bacterial quorum sensing inhibitors: Attractive alternatives for control of infectious pathogens showing multiple drug resistance. Recent Pat. Antiinfect. Drug Discov. 2013, 8, 68–83. [Google Scholar] [CrossRef]
- Manefield, M.; de Nys, R.; Naresh, K.; Roger, R.; Givskov, M.; Peter, S.; Kjelleberg, S. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 1999, 145, 283–291. [Google Scholar] [CrossRef]
- Lin, Y.H.; Xu, J.L.; Hu, J.; Wang, L.H.; Ong, S.L.; Leadbetter, J.R.; Zhang, L.H. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 2003, 47, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Chhabra, S.R.; Cámara, M.; Williams, P.; Oger, P.; Dessaux, Y. N-acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology 2005, 151, 3313–3322. [Google Scholar] [CrossRef]
- Fetzner, S. Quorum quenching enzymes. J. Biotechnol. 2015, 201, 2–14. [Google Scholar] [CrossRef]
- Stevens, A.M.; Queneau, Y.; Soulere, L.; von Bodman, S.; Doutheau, A. Mechanisms and synthetic modulators of AHL-dependent gene regulation. Chem. Rev. 2011, 111, 4–27. [Google Scholar] [CrossRef] [PubMed]
- LaSarre, B.; Federle, M.J. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 2013, 77, 73–111. [Google Scholar] [CrossRef]
- Beury-Cirou, A.; Tannières, M.; Minard, C.; Soulère, L.; Rasamiravaka, T.; Dodd, R.H.; Queneau, Y.; Dessaux, Y.; Guillou, C.; Vandeputte, O.M.; et al. At a supra-physiological concentration, human sexual hormones act as quorum-sensing inhibitors. PLoS ONE 2013, 8, e83564. [Google Scholar] [CrossRef]
- Muh, U.; Schuster, M.; Heim, R.; Singh, A.; Olson, E.R.; Greenberg, E.P. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob. Agents Chemother. 2006, 50, 3674–3679. [Google Scholar] [CrossRef]
- Borlee, B.R.; Geske, G.D.; Blackwell, H.E.; Handelsman, J. Identification of synthetic inducers and inhibitors of the quorum-sensing regulator lasR in Pseudomonas aeruginosa by high-throughput screening. Appl. Environ. Microbiol. 2010, 76, 8255–8258. [Google Scholar] [CrossRef] [PubMed]
- Götz, C.; Fekete, A.; Gebefuegi, I.; Forczek, S.T.; Fuksová, K.; Li, X.; Englmann, M.; Gryndler, M.; Hartmann, A.; Matucha, M.; et al. Uptake, degradation and chiral discrimination of N-acyl-d/l-homoserine lactones by barley (Hordeum vulgare) and yam bean (Pachyrhizus erosus) plants. Anal. Bioanal. Chem. 2007, 389, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Adonizio, A.L.; Downum, K.; Bennett, B.C.; Mathee, K. Anti-quorum sensing activity of medicinal plants in southern Florida. J. Ethnopharmacol. 2006, 105, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Bouyahya, A.; Dakka, N.; Et-Touys, A.; Abrini, J.; Bakri, Y. Medicinal plant products targeting quorum sensing for combating bacterial infections. Asian Pac. J. Trop. Med. 2017, 10, 729–743. [Google Scholar] [CrossRef] [PubMed]
- Ahumedo, M.; Díaz, A.; Vivas-Reyes, R. Theoretical and structural analysis of the active site of the transcriptional regulators LasR and TraR, using molecular docking methodology for identifying potential analogues of acyl homoserine lactones (AHLs) with anti-quorum sensing activity. Eur. J. Med. Chem. 2010, 45, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Soulère, L.; Sabbah, M.; Fontaine, F.; Queneau, Y.; Doutheau, A. LuxR-dependent quorum sensing: Computer aided discovery of new inhibitors structurally unrelated to N-acylhomoserine lactones. Bioorg. Med. Chem. Lett. 2010, 20, 4355–4358. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Oger, P.M.; Chapelle, E.; Adeline, M.T.; Faure, D.; Dessaux, Y. A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Appl. Environ. Microbiol. 2008, 74, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Rubio-Portillo, E.; Antón, J.; Ramos-Esplá, A.A.; Quesada, E.; Llamas, I. Selection of the N-acylhomoserine lactone-degrading bacterium Alteromonas stellipolaris PQQ-42 and of its potential for biocontrol in aquaculture. Front. Microbiol. 2016, 7, 646. [Google Scholar] [CrossRef] [PubMed]
- Reina, J.C.; Torres, M.; Llamas, I. Stenotrophomonas maltophilia AHL-degrading strains isolated from marine invertebrate microbiota attenuate the virulence of Pectobacterium carotovorum and Vibrio corallilyticus. Mar. Biotechnol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Romero, M.; Prado, S.; Dubert, J.; Tahrioui, A.; Otero, A.; Llamas, I. N-acylhomoserine lactone-degrading bacteria isolated from hatchery bivalve larval cultures. Microbiol. Res. 2013, 168, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Williamson, L.L.; Borlee, B.R.; Schloss, P.D.; Guan, C.; Allen, H.K.; Handelsman, J. Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl. Environ. Microbiol. 2005, 71, 6335–6344. [Google Scholar] [CrossRef]
- Riaz, K.; Elmerich, C.; Moreira, D.; Raffoux, A.; Dessaux, Y.; Faure, D. A metagenomic analysis of soil bacteria extends the diversity of quorum-quenching lactonases. Environ. Microbiol. 2008, 10, 560–570. [Google Scholar] [CrossRef]
- Bijtenhoorn, P.; Schipper, C.; Hornung, C.; Quitschau, M.; Grond, S.; Weiland, N.; Streit, W.R. BpiB05, a novel metagenome-derived hydrolase acting on N-acylhomoserine lactones. J. Biotechnol. 2011, 155, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Schipper, C.; Hornung, C.; Bijtenhoorn, P.; Quitschau, M.; Grond, S.; Streit, W.R. Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2009, 75, 224–233. [Google Scholar] [CrossRef]
- Romero, M.; Martin-Cuadrado, A.; Otero, A. Determination of whether quorum quenching is a common activity in marine bacteria by analysis of cultivable bacteria and metagenomic sequences. Appl. Environ. Microbiol. 2012, 78, 6345–6348. [Google Scholar] [CrossRef]
- Tannières, M.; Beury-Cirou, A.; Vigouroux, A.; Mondy, S.; Pellissier, F.; Dessaux, Y.; Faure, D. A Metagenomic study highlights phylogenetic proximity of quorum-quenching and xenobiotic-degrading amidases of the AS-family. PLoS ONE 2013, 8, e65473. [Google Scholar] [CrossRef]
- Weiland-Bräuer, N.; Kisch, M.J.; Pinnow, N.; Liese, A.; Schmitz, R.A. Highly effective inhibition of biofilm formation by the first metagenome-derived AI-2 quenching enzyme. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Yaniv, K.; Golberg, K.; Kramarsky-Winter, E.; Marks, R.; Pushkarev, A.; Béjà, O.; Kushmaro, A. Functional marine metagenomic screening for anti-quorum sensing and anti-biofilm activity. Biofouling 2017, 33, 1–13. [Google Scholar] [CrossRef]
- Torres, M.; Uroz, S.; Salto, R.; Fauchery, L.; Quesada, E.; Llamas, I. HqiA, a novel quorum-quenching enzyme which expands the AHL lactonase family. Sci. Rep. 2017, 7, 943. [Google Scholar] [CrossRef]
- Shinohara, M.; Nakajima, N.; Uehara, Y. Purification and characterization of a novel esterase (β-hydroxypalmitate methyl ester hydrolase) and prevention of the expression of virulence by Ralstonia solanacearum. J. Appl. Microbiol. 2007, 103, 152–162. [Google Scholar] [CrossRef]
- Pustelny, C.; Albers, A.; Büldt-Karentzopoulos, K.; Parschat, K.; Chhabra, S.R.; Cámara, M.; Williams, P.; Fetzner, S. Dioxygenase-mediated quenching of quinolone-dependent quorum sensing in Pseudomonas aeruginosa. Chem. Biol. 2009, 16, 1259–1267. [Google Scholar] [CrossRef] [PubMed]
- Newman, K.L.; Chatterjee, S.; Ho, K.A.; Lindow, S.E. Virulence of plant pathogenic bacteria attenuated by degradation of fatty acid cell-to-cell signaling Factors. Mol. Plant-Microbe Interact. 2008, 21, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kim, Y.W.; Lee, S.; Lee, S.H.; Nahm, C.H.; Kwon, H.; Park, P.K.; Choo, K.H.; Koyuncu, I.; Drews, A.; et al. Stopping autoinducer-2 chatter by means of an indigenous bacterium (Acinetobacter sp. DKY-1): a new antibiofouling strategy in a membrane bioreactor for wastewater treatment. Environ. Sci. Technol. 2018, 52, 6237–6245. [Google Scholar] [CrossRef]
- Swift, S.; Lynch, M.J.; Fish, L.; Kirke, D.F.; Tomás, J.M.; Stewart, G.S.A.B.; Williams, P.; Tomas, J.M.; Stewart, G.S.A.B.; Williams, P. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect. Immun. 1999, 67, 5192–5199. [Google Scholar]
- Swift, S.; Karlyshev, A.V.; Fish, L.; Durant, E.L.; Winson, M.K.; Chhabra, S.R.; Williams, P.; Macintyre, S.; Stewart, G.S. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 1997, 179, 5271–5281. [Google Scholar] [CrossRef]
- Lynch, M.J.; Swift, S.; Kirke, D.F.; Keevil, C.W.; Dodd, C.E.R.; Williams, P. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 2002, 4, 18–28. [Google Scholar] [CrossRef]
- Chen, R.; Zhou, Z.; Cao, Y.; Bai, Y.; Yao, B. High yield expression of an AHL-lactonase from Bacillus sp. B546 in Pichia pastoris and its application to reduce Aeromonas hydrophila mortality in aquaculture. Microb. Cell Fact. 2010, 9, 39. [Google Scholar] [CrossRef]
- Cao, Y.; He, S.; Zhou, Z.; Zhang, M.; Mao, W.; Zhang, H.; Yaoa, B. Orally administered thermostable N-acyl homoserine lactonase from Bacillus sp. strain AI96 attenuates Aeromonas hydrophila infection in zebrafish. Appl. Environ. Microbiol. 2012, 78, 1899–1908. [Google Scholar] [CrossRef]
- Zhou, S.; Zhang, A.; Yin, H.; Chu, W. Bacillus sp. QSI-1 Modulate quorum sensing signals reduce Aeromonas hydrophila level and alter gut microbial community structure in fish. Front. Cell. Infect. Microbiol. 2016, 6, 1–8. [Google Scholar] [CrossRef]
- Ponnusamy, K.; Paul, D.; Kweon, J.H. Inhibition of quorum sensing mechanism and Aeromonas hydrophila biofilm formation by vanillin. Environ. Eng. Sci. 2009, 26, 1359–1363. [Google Scholar] [CrossRef]
- Husain, F.M.; Ahmad, I.; Khan, M.S.; Al-Shabib, N.A. Trigonella foenum-graceum (seed) extract interferes with quorum sensing regulated traits and biofilm formation in the strains of Pseudomonas aeruginosa and Aeromonas hydrophila. Evid.-Based Complement. Altern. Med. 2015, 2015, 1–10. [Google Scholar] [CrossRef]
- Meng, L.; Du, Y.; Liu, P.; Li, X.; Liu, Y. Involvement of LuxS in Aeromonas salmonicida metabolism, virulence and infection in Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 2017, 64, 260–269. [Google Scholar] [CrossRef]
- Rasch, M.; Kastbjerg, V.; Bruhn, J.; Dalsgaard, I.; Givskov, M.; Gram, L. Quorum sensing signals are produced by Aeromonas salmonicida and quorum sensing inhibitors can reduce production of a potential virulence factor. Dis. Aquat. Organ. 2007, 78, 105–113. [Google Scholar] [CrossRef]
- Lupp, C.; Ruby, E.G. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization. J. Bacteriol. 2005, 187, 3620–3629. [Google Scholar] [CrossRef]
- Han, Y.; Li, X.; Qi, Z.; Zhang, X.H.; Bossier, P. Detection of different quorum-sensing signal molecules in a virulent Edwardsiella tarda strain LTB-4. J. Appl. Microbiol. 2010, 108, 139–147. [Google Scholar] [CrossRef]
- Zhang, M.; Sun, K.; Sun, L. Regulation of autoinducer 2 production and luxS expression in a pathogenic Edwardsiella tarda strain. Microbiology 2008, 154, 2060–2069. [Google Scholar] [CrossRef]
- Romero, M.; Muras, A.; Mayer, C.; Buján, N.; Magariños, B.; Otero, A. In vitro quenching of fish pathogen Edwardsiella tarda AHL production using marine bacterium Tenacibaculum sp. strain 20-J cell extracts. Dis. Aquat. Organ. 2014, 108, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jiao, X.D.; Hu, Y.H.; Sun, L. Attenuation of Edwardsiella tarda virulence by small peptides that interfere with luxS/autoinducer type 2 quorum sensing. Appl. Environ. Microbiol. 2009, 75, 3882–3890. [Google Scholar] [CrossRef]
- Ye, J.; Ma, Y.; Liu, Q.; Zhao, D.L.; Wang, Q.Y.; Zhang, Y.X. Regulation of Vibrio alginolyticus virulence by the LuxS quorum-sensing system. J. Fish Dis. 2008, 31, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Rasch, M.; Buch, C.; Austin, B.; Slierendrecht, W.; Ekmann, K.; Larsen, J.; Johansen, C.; Riedel, K.; Eberl, L.; Givskov, M.; et al. An inhibitor of bacterial quorum sensing reduces mortalities caused by vibriosis rainbow trout (Oncorthynchus inmykiss, Walbaum). Syst. Appl. Microbiol. 2004, 27, 350–359. [Google Scholar] [CrossRef]
- Brackman, G.; Celen, S.; Hillaert, U.; Van Calenbergh, S.; Cos, P.; Maes, L.; Nelis, H.J.; Coenye, T. Structure-activity relationship of cinnamaldehyde analogs as inhibitors of AI-2 based quorum sensing and their effect on virulence of Vibrio spp. PLoS ONE 2011, 6, e16084. [Google Scholar] [CrossRef]
- Haldar, S.; Chatterjee, S.; Sugimoto, N.; Das, S.; Chowdhury, N.; Hinenoya, A.; Asakura, M.; Yamasaki, S. Identification of Vibrio campbellii isolated from diseased farm-shrimps from south India and establishment of its pathogenic potential in an Artemia model. Microbiology 2011, 157, 179–188. [Google Scholar] [CrossRef]
- Noor, N.M.; Defoirdt, T.; Alipiah, N.; Karim, M.; Daud, H.; Natrah, I. Quorum sensing is required for full virulence of Vibrio campbellii towards tiger grouper (Epinephelus fuscoguttatus) larvae. J. Fish Dis. 2019. [Google Scholar] [CrossRef]
- Defoirdt, T.; Crab, R.; Wood, T.K.; Sorgeloos, P.; Verstraete, W.; Bossier, P. Quorum sensing-disrupting brominated furanones protect the gnotobiotic brine shrimp Artemia franciscana from pathogenic Vibrio harveyi, Vibrio campbellii and Vibrio parahaemolyticus isolates. Appl. Environ. Microbiol. 2006, 72, 6419–6423. [Google Scholar] [CrossRef]
- Tait, K.; Hutchison, Z.; Thompson, F.L.; Munn, C.B. Quorum sensing signal production and inhibition by coral-associated vibrios. Environ. Microbiol. Rep. 2010, 2, 145–150. [Google Scholar] [CrossRef]
- Bassler, B.L.; Wright, M.; Showalter, R.E.; Silverman, M.R. Intercellular signalling in Vibrio harveyi: Sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 1993, 9, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.G.; Meighen, E.A. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 1989, 264, 21670–21676. [Google Scholar]
- Santhakumari, S.; Jayakumar, R.; Logalakshmi, R.; Prabhu, N.M.; Abdul Nazar, A.K.; Karutha Pandian, S.; Veera Ravi, A. In vitro and in vivo effect of 2,6-di-tert-butyl-4-methylphenol as an antibiofilm agent against quorum sensing mediated biofilm formation of Vibrio spp. Int. J. Food Microbiol. 2018, 281, 60–71. [Google Scholar] [CrossRef]
- Ni, N.; Choudhary, G.; Li, M.; Wang, B. Pyrogallol and its analogs can antagonize bacterial quorum sensing in Vibrio harveyi. Bioorg. Med. Chem. Lett. 2008, 18, 1567–1572. [Google Scholar] [CrossRef]
- Henke, J.M.; Bassler, B.L. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 2004, 186, 3794–3805. [Google Scholar] [CrossRef]
- Bai, F.; Han, Y.; Chen, J.; Zhang, X.H. Disruption of quorum sensing in Vibrio harveyi by the AiiA protein of Bacillus thuringiensis. Aquaculture 2008, 274, 36–40. [Google Scholar] [CrossRef]
- Pande, G.S.J.; Scheie, A.A.; Benneche, T.; Wille, M.; Sorgeloos, P.; Bossier, P.; Defoirdt, T. Quorum sensing-disrupting compounds protect larvae of the giant freshwater prawn Macrobrachium rosenbergii from Vibrio harveyi infection. Aquaculture 2013, 406–407, 121–124. [Google Scholar] [CrossRef]
- Lowery, C.A.; Abe, T.; Park, J.; Eubanks, L.M.; Sawada, D.; Kaufmann, G.F.; Janda, K.D. Revisiting AI-2 quorum sensing inhibitors: Direct comparison of alkyl-DPD analogues and a natural product fimbrolide. J. Am. Chem. Soc. 2009, 131, 15584–15585. [Google Scholar] [CrossRef]
- Torres, M.; Hong, K.W.; Chong, T.M.; Reina, J.C.; Chan, K.G.; Dessaux, Y.; Llamas, I. Genomic analyses of two Alteromonas stellipolaris strains reveal traits with potential biotechnological applications. Sci. Rep. 2019, 9, 1215. [Google Scholar] [CrossRef]
- Valiente, E.; Bruhn, J.B.; Nielsen, K.F.; Larsen, J.L.; Roig, F.J.; Gram, L.; Amaro, C. Vibrio vulnificus produces quorum sensing signals of the AHL-class: Research article. FEMS Microbiol. Ecol. 2009, 69, 16–26. [Google Scholar] [CrossRef]
- Piper, K.R.; von Bodman, S.B.; Farrand, S.K. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 1993, 362, 448–450. [Google Scholar] [CrossRef]
- Zhang, L.; Murphy, P.J.; Kerr, A.; Tate, M.E. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature 1993, 362, 446–448. [Google Scholar] [CrossRef]
- Mhedbi-Hajri, N.; Yahiaoui, N.; Mondy, S.; Hue, N.; Pélissier, F.; Faure, D.; Dessaux, Y. Transcriptome analysis revealed that a quorum sensing system regulates the transfer of the pAt megaplasmid in Agrobacterium tumefaciens. BMC Genom. 2016, 17, 661. [Google Scholar] [CrossRef]
- Zhang, X.H.; Wang, L.; Zhang, L.H. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 2002, 99, 4638–4643. [Google Scholar] [CrossRef] [Green Version]
- Carlier, A.; Chevrot, R.; Dessaux, Y.; Faure, D. The assimilation of γ-butyrolactone in Agrobacterium tumefaciens C58 interferes with the accumulation of the N-acyl-homoserine lactone signal. Mol. Plant-Microbe Interact. 2004, 17, 951–957. [Google Scholar] [CrossRef]
- Haudecoeur, E.; Tannières, M.; Cirou, A.; Raffoux, A.; Dessaux, Y.; Faure, D. Different regulation and roles of lactonases AiiB and AttM in Agrobacterium tumefaciens C58. Mol. Plant-Microbe Interact. 2009, 22, 529–537. [Google Scholar] [CrossRef]
- McInnis, C.E.; Blackwell, H.E. Thiolactone modulators of quorum sensing revealed through library design and screening. Bioorg. Med. Chem. 2011, 19, 4820–4828. [Google Scholar] [CrossRef] [Green Version]
- Chernin, L.; Toklikishvili, N.; Ovadis, M.; Kim, S.; Ben-Ari, J.; Khmel, I.; Vainstein, A. Quorum-sensing quenching by rhizobacterial volatiles. Environ. Microbiol. Rep. 2011, 3, 698–704. [Google Scholar] [CrossRef]
- Liu, H.B.; Koh, K.P.; Kim, J.S.; Seo, Y.; Park, S. The effects of betonicine, floridoside, and isethionic acid from the red alga Ahnfeltiopsis flabelliformis on quorum-sensing activity. Biotechnol. Bioprocess Eng. 2008, 13, 458–463. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.G.; Kang, Y.; Jang, J.Y.; Jog, G.J.; Lim, J.Y.; Kim, S.; Suga, H.; Nagamatsu, T.; Hwang, I. Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol. Microbiol. 2004, 54, 921–934. [Google Scholar] [CrossRef]
- Kim, J.; Kang, Y.; Choi, O.; Jeong, Y.; Jeong, J.E.; Lim, J.Y.; Kim, M.; Moon, J.S.; Suga, H.; Hwang, I. Regulation of polar flagellum genes is mediated by quorum sensing and FlhDC in Burkholderia glumae. Mol. Microbiol. 2007, 64, 165–179. [Google Scholar] [CrossRef]
- Devescovi, G.; Bigirimana, J.; Degrassi, G.; Cabrio, L.; LiPuma, J.J.; Kim, J.; Hwang, I.; Venturi, V. Involvement of a quorum-sensing-regulated lipase secreted by a clinical isolate of Burkholderia glumae in severe disease symptoms in rice. Appl. Environ. Microbiol. 2007, 73, 4950–4958. [Google Scholar] [CrossRef]
- Kang, Y.; Goo, E.; Kim, J.; Hwang, I. Critical role of quorum sensing-dependent glutamate metabolism in homeostatic osmolality and outer membrane vesiculation in Burkholderia glumae. Sci. Rep. 2017, 7, 44195. [Google Scholar] [CrossRef]
- Cho, H.S.; Park, S.Y.; Ryu, C.M.; Kim, J.F.; Kim, J.G.; Park, S.H. Interference of quorum sensing and virulence of the rice pathogen Burkholderia glumae by an engineered endophytic bacterium. FEMS Microbiol. Ecol. 2007, 60, 14–23. [Google Scholar] [CrossRef]
- Chung, J.; Goo, E.; Yu, S.; Choi, O.; Lee, J.; Kim, J.; Kim, H.; Igarashi, J.; Suga, H.; Moon, J.S.; et al. Small-molecule inhibitor binding to an N-acyl-homoserine lactone synthase. Proc. Natl. Acad. Sci. USA 2011, 108, 12089–12094. [Google Scholar] [CrossRef]
- Nasser, W.; Bouillant, M.L.; Salmond, G.; Reverchon, S. Characterization of the Erwinia chrysanthemi expI-expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Mol. Microbiol. 1998, 29, 1391–1405. [Google Scholar] [CrossRef]
- Hussain, M.B.B.M.; Zhang, H.B.; Xu, J.L.; Liu, Q.; Jiang, Z.; Zhang, L.H. The acyl-homoserine lactone-type quorum-sensing system modulates cell motility and virulence of Erwinia chrysanthemi pv. zeae. J. Bacteriol. 2008, 190, 1045–1053. [Google Scholar] [CrossRef]
- Hosseinzadeh, S.; Shams-Bakhsh, M.; Yamchi, A. A comparative study on effect of two different aiiA genes on pathogenicity factors of Dickeya chrysanthemi pv. chrysanthemi. Arch. Phytopathol. Plant Prot. 2013, 46, 1468–1479. [Google Scholar] [CrossRef]
- Hosseinzadeh, S.; Shams-Bakhsh, M.; Sadeghizadeh, M. Attenuation and quantitation of virulence gene expression in quorum-quenched Dickeya chrysanthemi. Arch. Microbiol. 2017, 199, 51–61. [Google Scholar] [CrossRef]
- Nasser, W.; Dorel, C.; Wawrzyniak, J.; Van Gijsegem, F.; Groleau, M.C.; Déziel, E.; Reverchon, S. Vfm a new quorum sensing system controls the virulence of Dickeya dadantii. Environ. Microbiol. 2013, 15, 865–880. [Google Scholar] [CrossRef]
- Potrykus, M.; Golanowska, M.; Hugouvieux-Cotte-Pattat, N.; Lojkowska, E. Regulators involved in Dickeya solani virulence, genetic conservation and functional variability. Mol. Plant-Microbe Interact. 2015, 2015, 5–16. [Google Scholar] [CrossRef]
- Venturi, V.; Venuti, C.; Devescovi, G.; Lucchese, C.; Friscina, A.; Degrassi, G.; Aguilar, C.; Mazzucchi, U. The plant pathogen Erwinia amylovora produces acyl-homoserine lactone signal molecules in vitro and in planta. FEMS Microbiol. Lett. 2004, 241, 179–183. [Google Scholar] [CrossRef]
- Gao, Y.; Song, J.; Hu, B.; Zhang, L.; Liu, Q.; Liu, F. The luxS gene is involved in AI-2 production, pathogenicity, and some phenotypes in Erwinia amylovora. Curr. Microbiol. 2009, 58, 1–10. [Google Scholar] [CrossRef]
- Molina, L.; Rezzonico, F.; Defago, G.; Duffy, B. Autoinduction in Erwinia amylovora: Evidence of an acyl-homoserine lactone signal in the fire blight pathogen. J. Bacteriol. 2005, 187, 3206–3213. [Google Scholar] [CrossRef] [PubMed]
- von Bodman, S.B.; Majerczak, D.R.; Coplin, D.L. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc. Natl. Acad. Sci. USA 1998, 95, 7687–7692. [Google Scholar] [CrossRef] [PubMed]
- Koutsoudis, M.D.; Tsaltas, D.; Minogue, T.D.; von Bodman, S.B. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. USA 2006, 103, 5983–5988. [Google Scholar] [CrossRef]
- Oh, H.S.H.; Tan, C.H.; Low, J.H.J.; Rzechowicz, M.; Siddiqui, M.M.F.; Winters, H.; Kjelleberg, S.; Fane, A.G.; Rice, S.A. Quorum quenching bacteria can be used to inhibit the biofouling of reverse osmosis membranes. Water Res. 2017, 112, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Smadja, B.; Latour, X.; Faure, D.; Chevalier, S.; Dessaux, Y.; Orange, N. Involvement of N-acylhomoserine lactones throughout plant infection by Erwinia carotovora subsp. atroseptica (Pectobacterium atrosepticum). Mol. Plant. Microbe. Interact. 2004, 17, 1269–1278. [Google Scholar] [CrossRef]
- Liu, H.; Coulthurst, S.J.; Pritchard, L.; Hedley, P.E.; Ravensdale, M.; Humphris, S.; Burr, T.; Takle, G.; Brurberg, M.B.; Birch, P.R.J.; et al. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 2008, 4, e1000093. [Google Scholar] [CrossRef]
- Monson, R.; Burr, T.; Carlton, T.; Liu, H.; Hedley, P.; Toth, I.; Salmond, G.P.C. Identification of genes in the VirR regulon of Pectobacterium atrosepticum and characterization of their roles in quorum sensing-dependent virulence. Environ. Microbiol. 2013, 15, 687–701. [Google Scholar] [CrossRef] [PubMed]
- Bowden, S.D.; Hale, N.; Chung, J.C.S.; Hodgkinson, J.T.; Spring, D.R.; Welch, M. Surface swarming motility by Pectobacterium atrosepticum is a latent phenotype that requires O antigen and is regulated by quorum sensing. Microbiology 2013, 159, 2375–2385. [Google Scholar] [CrossRef]
- Barbey, C.; Crepin, A.; Bergeau, D.; Ouchiha, A.; Mijouin, L.; Taupin, L.; Orange, N.; Feuilloley, M.; Dufour, A.; Burini, J.F.; et al. In planta biocontrol of Pectobacterium atrosepticum by Rhodococcus erythropolis involves silencing of pathogen communication by the Rhodococcal gamma-lactone catabolic pathway. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Carlier, A.; Uroz, S.; Smadja, B.; Fray, R.; Latour, X.; Dessaux, Y.; Faure, D. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acyl homoserine lactonase activity. Appl. Environ. Microbiol. 2003, 69, 4989–4993. [Google Scholar] [CrossRef] [PubMed]
- Raoul des Essarts, Y.; Sabbah, M.; Comte, A.; Soulère, L.; Queneau, Y.; Dessaux, Y.; Hélias, V.; Faure, D.; des Essarts, Y.; Sabbah, M.; et al. N,N’-alkylated imidazolium-derivatives act as quorum-sensing inhibitors targeting the Pectobacterium atrosepticum-induced symptoms on potato tubers. Int. J. Mol. Sci. 2013, 14, 19976–19986. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Cui, Y.; Liu, Y.; Dumenyo, C.K.; Chatterjee, A.K. Global regulation in Erwinia species by Erwinia carotovora rsmA, a homologue of Escherichia coli csrA: Repression of secondary metabolites, pathogenicity and hypersensitive reaction. Microbiology 1996, 142, 427–434. [Google Scholar] [CrossRef]
- Cui, Y.; Chatterjee, A.; Liu, Y.; Dumenyo, C.K.; Chatterjee, A.K. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J. Bacteriol. 1995, 177, 5108–5115. [Google Scholar] [CrossRef]
- Liu, Y.; Cui, Y.; Mukherjee, A.; Chatterjee, A.K. Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol. Microbiol. 1998, 29, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Coulthurst, S.J.; Lilley, K.; Salmond, G. Genetic and proteomic analysis of the role of luxS in the enteric phytopathogen, Erwinia carotovora. Mol. Plant Pathol. 2006, 7, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Laasik, E.; Andresen, L.; Mäe, A. Type II quorum sensing regulates virulence in Erwinia carotovora ssp. carotovora. FEMS Microbiol. Lett. 2006, 258, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Manefield, M.; Welch, M.; Givskov, M.; Salmond, G.P.C.; Kjelleberg, S. Halogenated furanones from the red alga, Delisea pulchra, inhibit carbapenem antibiotic synthesis and exoenzyme virulence factor production in the phytopathogen Erwinia carotovora. FEMS Microbiol. Lett. 2001, 205, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, S.; Oh, T.; Oh, J.; Koo, B.; Yum, D.; Lee, J. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 2003, 149, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Colnaghi Simionato, A.V.; da Silva, D.S.; Lambais, M.R.; Carrilho, E. Characterization of a putative Xylella fastidiosa diffusible signal factor by HRGC-EI-MS. J. Mass Spectrom. 2007, 42, 1375–1381. [Google Scholar] [CrossRef]
- Chatterjee, S.; Wistrom, C.; Lindow, S.E. A cell-cell signaling sensor is required for virulence and insect transmission of Xylella fastidiosa. Proc. Natl. Acad. Sci. USA 2008, 105, 2670–2675. [Google Scholar] [CrossRef]
- Ionescu, M.; Zaini, P.A.; Baccari, C.; Tran, S.; da Silva, A.M.; Lindow, S.E. Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc. Natl. Acad. Sci. USA 2014, 111, E3910–E3918. [Google Scholar] [CrossRef]
- Kushner, D.; Kamekura, M. Physiology of halophilic bacteria. In Halophilic Bacteria; Rodriguez-Valera, F., Ed.; CRC Press: Boca Raton, FL, USA, 1988; Volume 1, pp. 109–138. [Google Scholar]
- Oren, A. Diversity of halophilic microorganisms: Environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biotechnol. 2002, 28, 56–63. [Google Scholar] [CrossRef]
- Ventosa, A. Unusual microorganisms from unusual habitats: Hypersaline environments. In Prokaryotic Diversity-Mechanism and Significance; Logan, N.A., Lappin-Scott, H.M., Oyston, P.C.F., Eds.; Cambridge University Press: Cambridge, UK, 2006; pp. 223–252. [Google Scholar]
- Margesin, R.; Schinner, F. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 2001, 5, 73–83. [Google Scholar] [CrossRef]
- Bhatnagar, I.; Kim, S.K. Immense essence of excellence: Marine microbial bioactive compounds. Mar. Drugs 2010, 8, 2673–2701. [Google Scholar] [CrossRef] [PubMed]
- Trincone, A. Marine biocatalysts: Enzymatic features and applications. Mar. Drugs 2011, 9, 478–499. [Google Scholar] [CrossRef]
- Hamza, F.; Zinjarde, S. Marine biodiversity as a resource for bioactive molecules as inhibitors of microbial quorum sensing phenotypes. In Biotechnological Applications of Quorum Sensing Inhibitors; Springer: Singapore, 2018; pp. 329–350. [Google Scholar]
- Llamas, I.; Amjres, H.; Mata, J.A.; Quesada, E.; Béjar, V. The Potential biotechnological applications of the exopolysaccharide produced by the halophilic bacterium Halomonas almeriensis. Molecules 2012, 17, 7103–7120. [Google Scholar] [CrossRef] [PubMed]
- Radchenkova, N.; Boyadzhieva, I.; Atanasova, N.; Poli, A.; Finore, I.; Di Donato, P.; Nicolaus, B.; Panchev, I.; Kuncheva, M.; Kambourova, M. Extracellular polymer substance synthesized by a halophilic bacterium Chromohalobacter canadensis 28. Appl. Microbiol. Biotechnol. 2018, 102, 4937–4949. [Google Scholar] [CrossRef]
- Chang, J.; Lee, Y.S.; Fang, S.J.; Park, I.H.; Choi, Y.L. Recombinant expression and characterization of an organic-solvent-tolerant α-amylase from Exiguobacterium sp. DAU5. Appl. Biochem. Biotechnol. 2013, 169, 1870–1883. [Google Scholar] [CrossRef]
- Scapin, S.M.N.; Souza, F.H.M.; Zanphorlin, L.M.; de Almeida, T.S.; Sade, Y.B.; Cardoso, A.M.; Pinheiro, G.L.; Murakami, M.T. Structure and function of a novel GH8 endoglucanase from the bacterial cellulose synthase complex of Raoultella ornithinolytica. PLoS ONE 2017, 12, e0176550. [Google Scholar] [CrossRef]
- Li, X.; Qian, P.; Wu, S.G.; Yu, H.Y. Characterization of an organic solvent-tolerant lipase from Idiomarina sp. W33 and its application for biodiesel production using Jatropha oil. Extremophiles 2014, 18, 171–178. [Google Scholar] [CrossRef]
- Houssen, W.E.; Jaspars, M. Isolation of Marine Natural Products. Methods Mol. Biol. 2012, 864, 367–392. [Google Scholar] [CrossRef]
- Nikapitiya, C. Bioactive secondary metabolites from marine microbes for drug discovery. Adv. Food Nutr. Res. 2012, 65, 363–387. [Google Scholar] [CrossRef]
- Teasdale, M.E.; Liu, J.; Wallace, J.; Akhlaghi, F.; Rowley, D.C. Secondary metabolites produced by the marine bacterium Halobacillus salinus that inhibit quorum sensing-controlled phenotypes in Gram-negative bacteria. Appl. Environ. Microbiol. 2009, 75, 567–572. [Google Scholar] [CrossRef]
- Teasdale, M.E.; Donovan, K.A.; Forschner-Dancause, S.R.; Rowley, D.C. Gram-positive marine bacteria as a potential resource for the discovery of quorum sensing inhibitors. Mar. Biotechnol. 2011, 13, 722–732. [Google Scholar] [CrossRef]
- D’Angelo-Picard, C.; Faure, D.; Penot, I.; Dessaux, Y. Diversity of N-acyl homoserine lactone-producing and -degrading bacteria in soil and tobacco rhizosphere. Environ. Microbiol. 2005, 7, 1796–1808. [Google Scholar] [CrossRef]
- Pierson, E.A.; Wood, D.; Cannon, J.; Blachere, F.; Leland, S. Interpopulation signaling via N-acyl-homoserine lactones among bacteria in the wheat rhizosphere. Mol. Plant-Microbe Interact. 1998, 11, 1078–1084. [Google Scholar] [CrossRef]
- Romero, M.; Martin-Cuadrado, A.B.; Roca-Rivada, A.; Cabello, A.M.; Otero, A. Quorum quenching in cultivable bacteria from dense marine coastal microbial communities. FEMS Microbiol. Ecol. 2011, 75, 205–217. [Google Scholar] [CrossRef]
- Muras, A.; López-Pérez, M.; Mayer, C.; Parga, A.; Amaro-Blanco, J.; Otero, A. High prevalence of quorum-sensing and quorum-quenching activity among cultivable bacteria and metagenomic sequences in the Mediterranean sea. Genes (Basel) 2018, 9, 100. [Google Scholar] [CrossRef]
- Natrah, F.; Defoirdt, T.; Sorgeloos, P.; Bossier, P. Disruption of bacterial cell-to-cell communication by marine organisms and its relevance to aquaculture. Mar. Biotechnol. 2011, 13, 109–126. [Google Scholar] [CrossRef]
- Saurav, K.; Costantino, V.; Venturi, V.; Steindler, L. Quorum sensing inhibitors from the sea discovered using bacterial N-acyl-homoserine lactone-based biosensors. Mar. Drugs 2017, 15, 53. [Google Scholar] [CrossRef]
- Chen, J.; Wang, B.; Lu, Y.; Guo, Y.; Sun, J.; Wei, B.; Zhang, H.; Wang, H. Quorum sensing inhibitors from marine microorganisms and their synthetic derivatives. Mar. Drugs 2019, 17, 80. [Google Scholar] [CrossRef]
- Natrah, F.M.I.; Kenmegne, M.M.; Wiyoto, W.; Sorgeloos, P.; Bossier, P.; Defoirdt, T. Effects of micro-algae commonly used in aquaculture on acyl-homoserine lactone quorum sensing. Aquaculture 2011, 317, 53–57. [Google Scholar] [CrossRef]
- Saurav, K.; Bar-Shalom, R.; Haber, M.; Burgsdorf, I.; Oliviero, G.; Costantino, V.; Morgenstern, D.; Steindler, L. In search of alternative antibiotic drugs: Quorum-quenching activity in sponges and their bacterial isolates. Front. Microbiol. 2016, 7, 1–18. [Google Scholar] [CrossRef]
- Martín-Rodríguez, A.; Reyes, F.; Martín, J.; Pérez-Yépez, J.; León-Barrios, M.; Couttolenc, A.; Espinoza, C.; Trigos, Á.; Martín, V.; Norte, M.; et al. Inhibition of bacterial quorum sensing by extracts from aquatic fungi: First report from marine endophytes. Mar. Drugs 2014, 12, 5503–5526. [Google Scholar] [CrossRef]
- Rehman, Z.U.; Leiknes, T. Quorum-quenching bacteria isolated from Red sea sediments reduce biofilm formation by Pseudomonas aeruginosa. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Gutiérrez-Barranquero, J.A.; Reen, F.J.; Parages, M.L.; McCarthy, R.; Dobson, A.D.W.; O’Gara, F. Disruption of N-acyl-homoserine lactone-specific signalling and virulence in clinical pathogens by marine sponge bacteria. Microb. Biotechnol. 2017. [Google Scholar] [CrossRef]
- Costantino, V.; Della Sala, G.; Saurav, K.; Teta, R.; Bar-Shalom, R.; Mangoni, A.; Steindler, L. Plakofuranolactone as a quorum quenching agent from the Indonesian sponge Plakortis cf. lita. Mar. Drugs 2017, 15, 59. [Google Scholar] [CrossRef]
- Blanchet, E.; Prado, S.; Stien, D.; Oliveira da Silva, J.; Ferandin, Y.; Batailler, N.; Intertaglia, L.; Escargueil, A.; Lami, R. Quorum sensing and quorum quenching in the Mediterranean seagrass Posidonia oceanica microbiota. Front. Mar. Sci. 2017, 4. [Google Scholar] [CrossRef]
- Pande, G.S.J.; Natrah, F.M.I.; Flandez, A.V.B.; Kumar, U.; Niu, Y.; Bossier, P.; Defoirdt, T. Isolation of AHL-degrading bacteria from micro-algal cultures and their impact on algal growth and on virulence of Vibrio campbellii to prawn larvae. Appl. Microbiol. Biotechnol. 2015, 99, 10805–10813. [Google Scholar] [CrossRef]
- Defoirdt, T.; Benneche, T.; Brackman, G.; Coenye, T.; Sorgeloos, P.; Scheie, A.A. A Quorum sensing-disrupting brominated thiophenone with a promising therapeutic potential to treat luminescent vibriosis. PLoS ONE 2012, 7, e41788. [Google Scholar] [CrossRef]
- Chang, H.; Zhou, J.; Zhu, X.; Yu, S.; Chen, L.; Jin, H.; Cai, Z. Strain identification and quorum sensing inhibition characterization of marine-derived Rhizobium sp. NAO1. R. Soc. Open Sci. 2017, 4, 170025. [Google Scholar] [CrossRef]
- Chen, X.; Chen, J.; Yan, Y.; Chen, S.; Xu, X.; Zhang, H.; Wang, H. Quorum sensing inhibitors from marine bacteria Oceanobacillus sp. XC22919. Nat. Prod. Res. 2018, 1–5. [Google Scholar] [CrossRef]
- Sun, S.; Dai, X.; Sun, J.; Bu, X.; Weng, C.; Li, H.; Zhu, H. A diketopiperazine factor from Rheinheimera aquimaris QSI02 exhibits anti-quorum sensing activity. Sci. Rep. 2016, 6, 39637. [Google Scholar] [CrossRef]
- Miao, L.; Xu, J.; Yao, Z.; Jiang, Y.; Zhou, H.; Jiang, W.; Dong, K. The anti-quorum sensing activity and bioactive substance of a marine derived Streptomyces. Biotechnol. Biotechnol. Equip. 2017, 31, 1007–1015. [Google Scholar] [CrossRef]
- Mireille Ayé, A.; Bonnin-Jusserand, M.; Brian-Jaisson, F.; Ortalo-Magné, A.; Culioli, G.; Koffi Nevry, R.; Rabah, N.; Blache, Y.; Molmeret, M. Modulation of violacein production and phenotypes associated with biofilm by exogenous quorum sensing N-acylhomoserine lactones in the marine bacterium Pseudoalteromonas ulvae TC14. Microbiology 2015, 161, 2039–2051. [Google Scholar] [CrossRef]
- Liaqat, I.; Bachmann, R.T.; Edyvean, R.G.J. Type 2 quorum sensing monitoring, inhibition and biofilm formation in marine microrganisms. Curr. Microbiol. 2014, 68, 342–351. [Google Scholar] [CrossRef]
- Ni, N.; Chou, H.T.; Wang, J.; Li, M.; Lu, C.D.; Tai, P.C.; Wang, B. Identification of boronic acids as antagonists of bacterial quorum sensing in Vibrio harveyi. Biochem. Biophys. Res. Commun. 2008, 369, 590–594. [Google Scholar] [CrossRef]
- Brackman, G.; Celen, S.; Baruah, K.; Bossier, P.; Van Calenberg, S.; Nelis, H.J.; Coenye, T.; Van Calenbergh, S.; Nelis, H.J.; Coenye, T. AI-2 quorum-sensing inhibitors affect the starvation response and reduce virulence in several Vibrio species, most likely by interfering with LuxPQ. Microbiology 2009, 155, 4114–4122. [Google Scholar] [CrossRef]
- Shen, G.; Rajan, R.; Zhu, J.; Bell, C.E.; Pei, D. Design and synthesis of substrate and intermediate analogue inhibitors of S-ribosylhomocysteinase. J. Med. Chem. 2006, 49, 3003–3011. [Google Scholar] [CrossRef]
- Malladi, V.L.A.; Sobczak, A.J.; Meyer, T.M.; Pei, D.; Wnuk, S.F. Inhibition of LuxS by S-ribosylhomocysteine analogues containing a [4-aza]ribose ring. Bioorg. Med. Chem. 2011, 19, 5507–5519. [Google Scholar] [CrossRef]
- Li, M.; Ni, N.; Chou, H.T.; Lu, C.D.; Tai, P.C.; Wang, B. Structure-based discovery and experimental verification of novel AI-2 quorum sensing inhibitors against Vibrio harveyi. ChemMedChem 2008, 3, 1242–1249. [Google Scholar] [CrossRef]
- Zhu, P.; Peng, H.; Ni, N.; Wang, B.; Li, M. Novel AI-2 quorum sensing inhibitors in Vibrio harveyi identified through structure-based virtual screening. Bioorg. Med. Chem. Lett. 2012, 22, 6413–6417. [Google Scholar] [CrossRef]
- Jiang, T.; Zhu, P.; Du, L.; Li, M. Identification of AI-2 quorum sensing inhibitors in Vibrio harveyi through structure-based virtual screening. Methods Mol. Biol. 2018, 1673, 353–362. [Google Scholar] [CrossRef]
- Liu, N.; Yu, M.; Zhao, Y.; Cheng, J.; An, K.; Zhang, X.H. PfmA, a novel quorum-quenching N-acylhomoserine lactone acylase from Pseudoalteromonas flavipulchra. Microbiology 2017, 163, 1389–1398. [Google Scholar] [CrossRef]
- Tang, K.; Su, Y.; Brackman, G.; Cui, F.; Zhang, Y.; Shi, X.; Coenye, T.; Zhang, X.H. MomL, a novel marine-derived N-acyl homoserine lactonase from Muricauda olearia. Appl. Environ. Microbiol. 2015, 81, 774–782. [Google Scholar] [CrossRef]
- Romero, M.; Avendaño-Herrera, R.; Magariños, B.; Cámara, M.; Otero, A. Acylhomoserine lactone production and degradation by the fish pathogen Tenacibaculum maritimum, a member of the Cytophaga-Flavobacterium-Bacteroidetes (CFB) group. FEMS Microbiol. Lett. 2010, 304, 131–139. [Google Scholar] [CrossRef]
- Cai, X.; Yu, M.; Shan, H.; Tian, X.; Zheng, Y.; Xue, C.; Zhang, X.H. Characterization of a novel N-acylhomoserine lactonase RmmL from Ruegeria mobilis YJ3. Mar. Drugs 2018, 16, 370. [Google Scholar] [CrossRef] [PubMed]
- See-Too, W.S.; Convey, P.; Pearce, D.A.; Chan, K.G. Characterization of a novel N-acylhomoserine lactonase, AidP, from Antarctic Planococcus sp. Microb. Cell Fact. 2018, 17, 179. [Google Scholar] [CrossRef]
- Zhou, S.; Xia, Y.; Zhu, C.; Chu, W. Isolation of marine Bacillus sp. with antagonistic and organic-substances-degrading activities and its potential application as a fish probiotic. Mar. Drugs 2018, 16, 196. [Google Scholar] [CrossRef]
- Morohoshi, T.; Nakazawa, S.; Ebata, A.; Kato, N.; Ikeda, T. Identification and characterization of N-acylhomoserine lactone-acylase from the fish intestinal Shewanella sp. strain MIB015. Biosci. Biotechnol. Biochem. 2008, 72, 1887–1893. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.; Diggle, S.P.; Heeb, S.; Cámara, M.; Otero, A. Quorum quenching activity in Anabaena sp. PCC 7120: identification of AiiC, a novel AHL-acylase. FEMS Microbiol. Lett. 2008, 280, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Romero, M.; Muras, A.; Otero, A. Aii20J, a wide-spectrum thermostable N-acylhomoserine lactonase from the marine bacterium Tenacibaculum sp. 20J, can quench AHL-mediated acid resistance in Escherichia coli. Appl. Microbiol. Biotechnol. 2015, 99, 9523–9539. [Google Scholar] [CrossRef] [PubMed]
- Yates, E.A.; Philipp, B.; Buckley, C.; Atkinson, S.; Chhabra, S.R.; Sockett, R.E.; Goldner, M.; Dessaux, Y.; Cámara, M.; Smith, H.; et al. n-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 2002, 70, 5635–5646. [Google Scholar] [CrossRef]
- Byers, J.; Lucas, C.; Salmond, G.; Welch, M. Nonenzymatic turnover of an Erwinia carotovora quorum-sensing signaling molecule. J. Biotechnol. 2002, 184, 1163–1171. [Google Scholar] [CrossRef]
- Ng, F.S.W.; Wright, D.M.; Seah, S.Y.K. Characterization of a phosphotriesterase-like lactonase from Sulfolobus solfataricus and its immobilization for disruption of quorum sensing. Appl. Environ. Microbiol. 2011, 77, 1181–1186. [Google Scholar] [CrossRef]
- Demirjian, D.C.; Morís-Varas, F.; Cassidy, C.S. Enzymes from extremophiles. Curr. Opin. Chem. Biol. 2001, 5, 144–151. [Google Scholar] [CrossRef]
- Ravot, G.; Masson, J.M.; Lefèvre, F. Applications of extremophiles: The industrial screening of extremophiles for valuable biomolecules. Methods Microbiol. 2006, 35, 785–813. [Google Scholar] [CrossRef]
- Llamas, I.; Quesada, E.; Martínez-Cánovas, M.J.; Gronquist, M.; Eberhard, A.; González, J.E. Quorum sensing in halophilic bacteria: detection of N-acyl-homoserine lactones in the exopolysaccharide-producing species of Halomonas. Extremophiles 2005, 9, 333–341. [Google Scholar] [CrossRef]
- Tahrioui, A.; Schwab, M.; Quesada, E.; Llamas, I. Quorum sensing in some representative species of Halomonadaceae. Life 2013, 3, 260–275. [Google Scholar] [CrossRef]
- Abbamondi, G.R.; Suner, S.; Cutignano, A.; Grauso, L.; Nicolaus, B.; Oner, E.T.; Tommonaro, G. Identification of N-hexadecanoyl-l-homoserine lactone (C16-AHL) as signal molecule in halophilic bacterium Halomonas smyrnensis AAD6. Ann. Microbiol. 2016, 66, 1329–1333. [Google Scholar] [CrossRef]
- Sewald, X.; Saum, S.; Palm, P.; Pfeiffer, F.; Oesterhelt, D.; Muller, V. Autoinducer-2-producing protein LuxS, a novel salt- and chloride-induced protein in the moderately halophilic bacterium Halobacillus halophilus. Appl. Environ. Microbiol. 2007, 73, 371–379. [Google Scholar] [CrossRef]
- Tommonaro, G.; Abbamondi Gennaro, R.; Toksoy Oner, R.; Nicolaus, B. Investigating the quorum sensing system in halophilic bacteria. Halophiles 2015, 189–207. [Google Scholar] [CrossRef]
- Tommonaro, G.; Abbamondi, G.R.; Iodice, C.; Tait, K.; De Rosa, S. Diketopiperazines produced by the halophilic archaeon, Haloterrigena hispanica, activate AHL bioreporters. Microb. Ecol. 2012, 63, 490–495. [Google Scholar] [CrossRef]
- Montgomery, K.; Charlesworth, J.C.; LeBard, R.; Visscher, P.T.; Burns, B.P. Quorum sensing in extreme environments. Life 2013, 3, 131–148. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Dobretsov, S.; Al-Fori, M.; Gunasekera, S.P.; Sudesh, K.; Paul, V.J. Quorum-sensing inhibitory compounds from extremophilic microorganisms isolated from a hypersaline cyanobacterial mat. J. Ind. Microbiol. Biotechnol. 2013, 40, 759–772. [Google Scholar] [CrossRef]
- Singh, V.K.; Mishra, A.; Jha, B. Anti-quorum sensing and anti-biofilm activity of delftia tsuruhatensis extract by attenuating the quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2017, 7. [Google Scholar] [CrossRef]
- Kannan, V.; Bastas, K.; Devi, R. Scientific and economic impact of plant pathogenic bacteria. In Sustainable Approaches to Controlling Plant Pathogenic Bacteria; Kannan, V.R., Bastas, K.K., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 369–392. [Google Scholar]
- FAO. The State of World Fisheries and Aquaculture: Contributing to Food Security and Nutrition for All; FAO: Rome, Italy, 2016; ISBN 978-92-5-109185-2. [Google Scholar]
- Martins, P.M.M.; Merfa, M.V.; Takita, M.A.; De Souza, A.A. Persistence in phytopathogenic bacteria: Do we know enough? Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Smith, P. Antimicrobial resistance in aquaculture. Rev. Sci. Tech. 2008, 27, 243–264. [Google Scholar] [CrossRef] [PubMed]
- Akinbowale, O.L.; Peng, H.; Barton, M.D. Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. J. Appl. Microbiol. 2006, 100, 1103–1113. [Google Scholar] [CrossRef] [Green Version]
- WHO. Report of a joint FAO/OIE/WHO Expert Consulation on Antimicrobial Use in Aquaculture and Antimicrobial Resistance, Seoul, Republic of Korea; WHO: Geneva, Switzerland, 2006; pp. 13–16. [Google Scholar]
- Watts, J.E.M.; Schreier, H.J.; Lanska, L.; Hale, M.S. The rising tide of antimicrobial resistance in aquaculture: Sources, sinks and solutions. Mar. Drugs 2017, 15, 158. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.; Acuña, L.; Otero, A. Patents on quorum quenching: interfering with bacterial communication as a strategy to fight infections. Recent Pat. Biotechnol. 2012, 6, 2–12. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, L.; Zhang, M.; Liu, H.; Lu, P.; Lin, K. Quorum sensing inhibitors: A patent review (2014–2018). Expert Opin. Ther. Pat. 2018, 1–17. [Google Scholar] [CrossRef]
- Chu, W.; Zhou, S.; Zhu, W.; Zhuang, X. Quorum quenching bacteria Bacillus sp. QSI-1 protect zebrafish (Danio rerio) from Aeromonas hydrophila infection. Sci. Rep. 2014, 4, 5446. [Google Scholar] [CrossRef]
- Nahn, D.; Cam, D.; Wille, M.; Defoirdt, T.; Bossier, P.; Sorgeloos, P. Quorum quenching bacteria protect Macrobrachium rosenbergii larvae from Vibrio harveyi infection. J. Appl. Microbiol. 2010, 109, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Tinh, N.T.N.; Dierckens, K.; Sorgeloos, P.; Bossier, P. A review of the functionality of probiotics in the larviculture food chain. Mar. Biotechnol. 2008, 10, 1–12. [Google Scholar] [CrossRef]
- Vinoj, G.; Vaseeharan, B.; Thomas, S.; Spiers, A.J.; Shanthi, S. Quorum-quenching activity of the AHL-lactonase from Bacillus licheniformis DAHB1 inhibits Vibrio biofilm formation in vitro and reduces shrimp intestinal colonisation and mortality. Mar. Biotechnol. 2014, 16, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Torabi Delshad, S.; Soltanian, S.; Sharifiyazdi, H.; Haghkhah, M.; Bossier, P. Identification of N-acyl homoserine lactone-degrading bacteria isolated from rainbow trout (Oncorhynchus mykiss). J. Appl. Microbiol. 2018, 1–2. [Google Scholar] [CrossRef]
- Weerasekara, N.A.; Choo, K.H.; Lee, C.H. Biofouling control: Bacterial quorum quenching versus chlorination in membrane bioreactors. Water Res. 2016, 103, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Dobretsov, S.; Teplitski, M.; Bayer, M.; Gunasekera, S.; Proksch, P.; Paul, V. Inhibition of marine biofouling by bacterial quorum sensing inhibitors. Biofouling 2011, 27, 893–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeon, K.M.; Lee, C.H.; Kim, J. Magnetic enzyme carrier for effective biofouling control in the membrane bioreactor based on enzymatic quorum quenching. Environ. Sci. Technol. 2009, 43, 7403–7409. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Choi, D.C.; Yeon, K.M.; Kim, S.R.; Lee, C.H. Enzyme-immobilized nanofiltration membrane to mitigate biofouling based on quorum quenching. Environ. Sci. Technol. 2011, 45, 1601–1607. [Google Scholar] [CrossRef]
- Lade, H.; Paul, D.; Kweon, J.H. Quorum quenching mediated approaches for control of membrane biofouling. Int. J. Biol. Sci. 2014, 10, 550–565. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.N.; Upreti, D.K.; Singh, B.R.; Defoirdt, T.; Gupta, V.K.; De Souza, A.O.; Singh, H.B.; Barreira, J.C.; Ferreira, I.C.; Vahabi, K. Bactericidal, quorum quenching and anti-biofilm nanofactories: A new niche for nanotechnologists. Crit. Rev. Biotechnol. 2016, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nahm, C.H.; Choi, D.C.; Kwon, H.; Lee, S.H.S.; Lee, S.H.S.; Lee, K.; Choo, K.H.; Lee, J.K.; Lee, C.H.; Park, P.K. Application of quorum quenching bacteria entrapping sheets to enhance biofouling control in a membrane bioreactor with a hollow fiber module. J. Memb. Sci. 2017, 526, 264–271. [Google Scholar] [CrossRef]
- Lee, J.; Lee, I.; Nam, J.; Hwang, D.S.; Yeon, K.M.; Kim, J. Immobilization and stabilization of acylase on carboxylated polyaniline nanofibers for highly effective antifouling application via quorum quenching. ACS Appl. Mater. Interfaces 2017, 9, 15424–15432. [Google Scholar] [CrossRef]
- Kim, T.H.; Lee, I.; Yeon, K.M.; Kim, J. Biocatalytic membrane with acylase stabilized on intact carbon nanotubes for effective antifouling via quorum quenching. J. Memb. Sci. 2018, 554, 357–365. [Google Scholar] [CrossRef]
- Schultz, M.P.; Bendick, J.A.; Holm, E.R.; Hertel, W.M. Economic impact of biofouling on a naval surface ship. Biofouling 2011, 27, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helman, Y.; Chernin, L. Silencing the mob: Disrupting quorum sensing as a means to fight plant disease. Mol. Plant Pathol. 2015, 16, 316–329. [Google Scholar] [CrossRef]
- Pirhonen, M.; Flego, D.; Heikinheimo, R.; Palva, E.T. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993, 12, 2467–2476. [Google Scholar] [CrossRef]
- Põllumaa, L.; Alamäe, T.; Mäe, A. Quorum sensing and expression of virulence in Pectobacteria. Sensors 2012, 12, 3327–3349. [Google Scholar] [CrossRef]
- Piqué, N.; Miñana-Galbis, D.; Merino, S.; Tomás, J. Virulence Factors of Erwinia amylovora: A Review. Int. J. Mol. Sci. 2015, 16, 12836–12854. [Google Scholar] [CrossRef] [Green Version]
- Naughton, L.M.; An, S.; Hwang, I.; Chou, S.H.; He, Y.Q.; Tang, J.L.; Ryan, R.P.; Dow, J.M. Functional and genomic insights into the pathogenesis of Burkholderia species to rice. Environ. Microbiol. 2016, 18, 780–790. [Google Scholar] [CrossRef]
- Kai, K. Bacterial quorum sensing in symbiotic and pathogenic relationships with hosts. Biosci. Biotechnol. Biochem. 2018, 82, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Molina, L.; Constantinescu, F.; Michel, L.; Reimmann, C.; Duffy, B.; Défago, G. Degradation of pathogen quorum-sensing molecules by soil bacteria: A preventive and curative biological control mechanism. FEMS Microbiol. Ecol. 2003, 45, 71–81. [Google Scholar] [CrossRef]
- Mori, Y.; Hosoi, Y.; Ishikawa, S.; Hayashi, K.; Asai, Y.; Ohnishi, H.; Shimatani, M.; Inoue, K.; Ikeda, K.; Nakayashiki, H.; et al. Ralfuranones contribute to mushroom-type biofilm formation by Ralstonia solanacearum strain OE1-1. Mol. Plant Pathol. 2018, 19, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Cirou, A.; Diallo, S.; Kurt, C.; Latour, X.; Faure, D. Growth promotion of quorum-quenching bacteria in the rhizosphere of Solanum tuberosum. Environ. Microbiol. 2007, 9, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Cazenave, A.; Llovel, W. Contemporary sea level rise. Ann. Rev. Mar. Sci. 2010, 2, 145–173. [Google Scholar] [CrossRef]
- Nguyen, T.T.X.; Woodroffe, C.D. Assessing relative vulnerability to sea-level rise in the western part of the Mekong River Delta in Vietnam. Sustain. Sci. 2016, 11, 645–659. [Google Scholar] [CrossRef]
- Hernández-Delgado, E.A. The emerging threats of climate change on tropical coastal ecosystem services, public health, local economies and livelihood sustainability of small islands: Cumulative impacts and synergies. Mar. Pollut. Bull. 2015, 101, 5–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daliakopoulos, I.N.; Tsanis, I.K.; Koutroulis, A.; Kourgialas, N.N.; Varouchakis, A.E.; Karatzas, G.P.; Ritsema, C.J. The threat of soil salinity: A European scale review. Sci. Total Environ. 2016, 573, 727–739. [Google Scholar] [CrossRef]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]

| Bacterium | QS Signal Molecules | QS-Regulated Phenotypes | References | Possible QS Disruptors | References | ||
|---|---|---|---|---|---|---|---|
| AHLs | Others | QQ Enzymes | QSI Compounds | ||||
| Of marine importance | |||||||
| Aeromonas hydrophila | C4-HSL, C6-HSL | AI-2 | Production of extracellular protease and biofilm formation | [120,121,122] | AiiA lactonase | Vanillin, plant extracts and caffeine | [123,124,125,126,127] |
| Aeromonas salmonicida | C4-HSL, C6-HSL, 3OC6-HSL, C10-HSL | AI-2 | Production of extracellular protease | [120,121,128] | - | Sulphur-containing AHL-analogues | [129] |
| Aliivibrio fischeri | 3OC6-HSL, C8-HSL | AI-2 | Bioluminescence | [130] | - | - | - |
| Aliivibrio salmonicida | C6-HSL, 3OC6-HSL | AI-2 | Biolumenescence and biofilm | [32,40] | - | - | - |
| Edwarsiella tarda | C4-HSL, C6-HSL, 3OC6-HSL, C7-HSL | AI-2 | Production of extracellular protein | [39,131,132] | Aii20J lactonase | Small peptides | [133,134] |
| Vibrio alginolyticus | 3OHC4-HSL, 3OC10-HSL, 3OHC14-HSL | AI-2 | Biofilm formation | [42,135] | - | - | - |
| Vibrio anguillarum | C6-HSL, 3OC10-HSL, 3OHC10-HSL | AI-2, CAI-1 | Biofilm formation, Production of metalloprotease and pigments | [46] | Aac-like acylase | Furanones; cinnamaldehyde analogs | [41,104,136,137] |
| Vibrio campbelli | 3OHC4-HSL | AI-2, CAI-1 | Production of metalloprotease, siderophores and chitinase A | [30,44,45,138,139] | Furanones | [140] | |
| Vibrio corallilyticus | C4-HSL 3OH,C10-HSL | AI-2 | Control of motility, production of hemolysin, caseinase, amylase and alkaline phosphatase | [47,141] | HqiA lactonase, QuiP-like acylase, AiiA lactonase, AttM lactonase | - | [47,105] |
| Vibrio harveyi | 3OHC4-HSL | AI-2, CAI-1 | Bioluminescence, type III secretion system, extracellular toxin, metalloprotease and siderophore | [48,142,143] | AiiA lactonase | Furanones; 2,6-di-tert-butyl-4-methylphenol; cinnamaldehyde analogs; pyrogallol and analogs, AI-2 analogs | [137,140,144,145,146,147,148,149] |
| Vibrio mediterranei | C4-HSL, C6-HSL, 3OHC12-HSL 3OC13-HSL | AI-2 | Control of motility, production of DNAse, and chitinase | [22,47] | HqiA lactonase, Aac-like acylase, AiiA lactonase, AttM lactonase | - | [47,104,150] |
| Vibrio owensii | C12-HSL, 3OHC12-HSL | - | Control of motility, production of hemolysin, amylase, DNAse, chitinase and phosphatase | [47] | HqiA lactonase, AiiA lactonase, AttM lactonase | - | [47] |
| Vibrio vulnificus | C4-HSL, 3OC6-HSL 3OHC6-HSL | AI-2 | Production of metalloprotease, exoprotease and hemolysin | [36,151] | - | 2,6-di-tert-butyl-4-methylphenol; cinnamaldehyde analogs | [137,144] |
| Of agricultural importance | |||||||
| Agrobacterium tumefaciens | 3OC8-HSL, 3OHC8-HSL | Virulence plasmid conjugation | [152,153,154] | AttM (BlcC) lactonase, AiiB lactonase | Floridoside, betonicine, isethionic acid, thiolactones, dimethyl disulfide, hordenine, estrone | [95,155,156,157,158,159,160] | |
| Burkholderia glumae | C6-HSL, C8-HSL | - | Production of the phytotoxin toxoflavin and lipase, biogenesis of flagella, control of internal osmolarity | [161,162,163,164] | AiiA lactonase | AHL- analog J8-C8 (d) | [165,166] |
| Dickeya dadantii | C6-HSL, 3OC6-HSL | - | Partial control of pectate lyase synthesis, control of motility and cell aggregation | [167,168] | AiiA lactonase | - | [169,170] |
| Dickeya solani | C6-HSL (a), C8-HSL | Unknown (Vfm system) | Partial control of the production of macerating exoenzymes | [171,172] | - | - | - |
| Erwinia amylovora | 3OC6-HSL, 3OHC6-HSL | AI-2 | Possible partial control of virulence | [173,174,175] | - | - | - |
| Pantoea stewartii | 3OC6-HSL | Production of exopolysaccharide Bacterial adhesion and biofilm formation | [176,177] | AiiO lactonase | - | [178] | |
| Pectobacterium atrosepticum | 3OC6-HSL, C8-HSL, 3OC8-HSL, C10-HSL | Production and secretion of macerating exoenzymes, production of harpin, control of motility | [179,180,181,182] | AttM (BlcC) lactonase, AiiB lactonase, AiiA lactonase, QsdA lactonase | N,N’-alkylated imidazolium-derivatives | [183,184,185] | |
| Pectobacterium carotovorum | C6-HSL, 3OC6-HSL, 3OC8-HSL | AI-2 (b) | Production of macerating exoenzymes and antibiotics | [186,187,188,189,190] | HqiA lactonase, QuiP-like acylase, AiiA lactonase, AhlD lactonase, QsdA lactonase, QlcA lactonase, QsdB amidohydrolase, unidentified oxidoreductase | Furanones, dimethyl disulfide | [70,74,91,103,105,108,112,115,159,191,192] |
| Ralstonia solanacearum | C6-HSL (c), C8-HSL | 3OH-PAME | Production of exopolysaccharide I and macerating exoenzymes | [13] | β-hydroxy-palmitate methyl ester hydrolase | - | [116] |
| Xanthomonas campestris | - | DSF (cis-11-methyl-2-dodecenoic acid) | Production of exopolysaccharide and exoenzymes | [16] | Degradation of DSF by unidentified bacterial activities | - | [118] |
| Xyllela fastidiosa | - | Xf-DSF (12-methyl-tetradecanoic acid) | Adhesin production, biofilm stability, insect transmission, production of outer membrane vesicules and attachment to plant vessel cells | [193,194,195] | - | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres, M.; Dessaux, Y.; Llamas, I. Saline Environments as a Source of Potential Quorum Sensing Disruptors to Control Bacterial Infections: A Review. Mar. Drugs 2019, 17, 191. https://doi.org/10.3390/md17030191
Torres M, Dessaux Y, Llamas I. Saline Environments as a Source of Potential Quorum Sensing Disruptors to Control Bacterial Infections: A Review. Marine Drugs. 2019; 17(3):191. https://doi.org/10.3390/md17030191
Chicago/Turabian StyleTorres, Marta, Yves Dessaux, and Inmaculada Llamas. 2019. "Saline Environments as a Source of Potential Quorum Sensing Disruptors to Control Bacterial Infections: A Review" Marine Drugs 17, no. 3: 191. https://doi.org/10.3390/md17030191
APA StyleTorres, M., Dessaux, Y., & Llamas, I. (2019). Saline Environments as a Source of Potential Quorum Sensing Disruptors to Control Bacterial Infections: A Review. Marine Drugs, 17(3), 191. https://doi.org/10.3390/md17030191







