Identification of the Actinomycin D Biosynthetic Pathway from Marine-Derived Streptomyces costaricanus SCSIO ZS0073
Abstract
:1. Introduction
2. Results
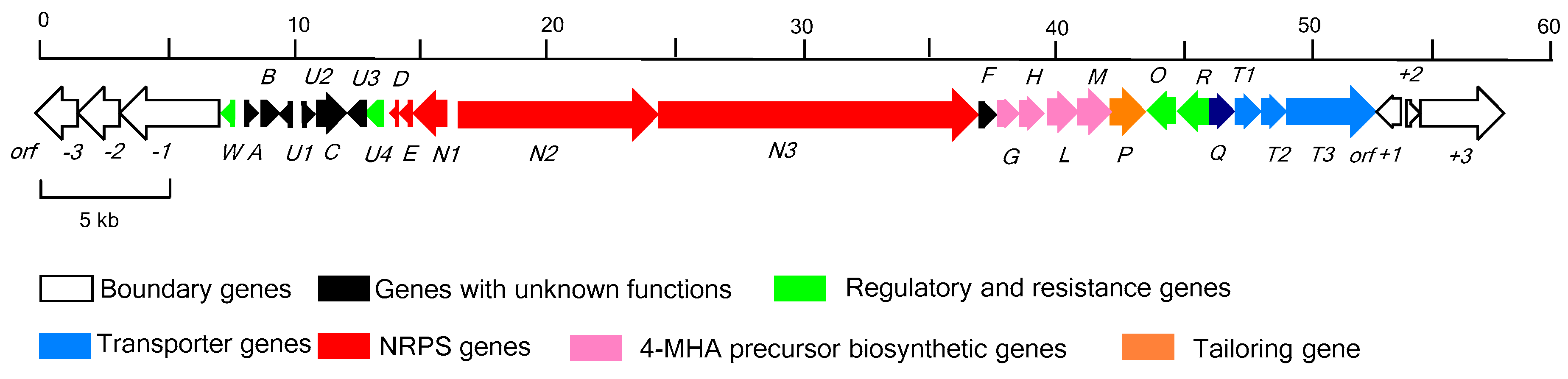
2.1. Identification of the Actinomycin D Biosynthetic Gene Cluster
2.2. Determination of the Actinomycin D Biosynthetic Gene Cluster Boundaries
2.3. NRPS Genes for Peptide Chain Assembly in Actinomycin D Biosynthesis
2.4. Biosynthetic Genes of 4-MHA
2.5. Regulatory and Self-Resistance Genes
2.6. Nonessential Genes of Unknown within the acn Cluster
2.7. Cytochrome P450 Gene acnP Is Responsible for the Hydroxylation of Proline in Actinomycin Xoβ
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Bacterial Strains, Plasmids and Culture Conditions
3.3. Whole Genome Scanning and Sequence Analysis
3.4. Genomic Library Construction and Screening
3.5. Inactivation of S. costaricanus SCSIO ZS0073 by λ-RED-Mediated PCR-Targeting Mutagenesis
3.6. Fermentation and Analysis of S. costaricanus SCSIO ZS0073 WT and Mutant Strains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bitzer, J.; Streibel, M.; Langer, H.J.; Grond, S. First Y-type actinomycins from Streptomyces with divergent structure-activity relationships for antibacterial and cytotoxic properties. Org. Biomol. Chem. 2009, 7, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Hollstein, U. Actinomycin. Chemistry and mechanism of action. Chem. Rev. 1974, 74, 625–652. [Google Scholar] [CrossRef]
- Cai, W.L.; Wang, X.; Elshahawi, S.I.; Ponomareva, L.V.; Liu, X.; McErlean, M.R.; Cui, Z.; Arlinghaus, A.L.; Thorson, J.S.; Van Lanen, S.G. Antibacterial and cytotoxic actinomycins Y6–Y9 and Zp from Streptomyces sp. strain Gö-GS12. J. Nat. Prod. 2016, 79, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, J.; Yuan, B.; Cao, C.; Qin, S.; Cao, X.; Bian, G.; Wang, Z.; Jiang, J. Methylated actinomycin D, a novel actinomycin D analog induces apoptosis in HepG2 cells through Fas- and mitochondria-mediated pathways. Mol. Carcinog. 2013, 52, 983–996. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Cao, P.; Ma, Y.T.; Luo, J.; Yan, Y.; Li, R.T.; Huang, S.X. A new actinomycin Z analogue with an additional oxygen bridge between chromophore and β-depsipentapeptide from Streptomyces sp. KIB-H714. Nat. Prod. Res. 2018, 2, 1–7. [Google Scholar] [CrossRef]
- Jiao, W.H.; Yuan, W.; Li, Z.Y.; Li, J.; Li, L.; Sun, J.B.; Gui, Y.H.; Wang, J.; Ye, B.P.; Lin, H.W. Anti-MRSA actinomycins D1–D4 from the marine sponge-associated Streptomyces sp. LHW52447. Tetrahedron 2018, 74, 5914–5919. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wang, M.; Tan, Y.; Hu, X.; He, H.; Xiao, C.L.; You, X.; Wang, Y.; Gan, M. Neo-actinomycins A and B, natural actinomycins bearing the 5 H-oxazolo [4-b] phenoxazine chromophore, from the marine-derived Streptomyces sp. IMB094. Sci. Rep. 2017, 7, 3591. [Google Scholar] [CrossRef]
- Bitzer, J.; Gesheva, V.; Zeeck, A. Actinomycins with altered threonine units in the β-peptidolactone. J. Nat. Prod. 2006, 69, 1153–1157. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.S.; Tseng, W.H.; Chuang, C.Y.; Hou, M.H. The structural basis of actinomycin D-binding induces nucleotide flipping out, a sharp bend and a left-handed twist in CGG triplet repeats. Nucleic Acids Res. 2013, 41, 4284–4294. [Google Scholar] [CrossRef]
- Das, T.; Nair, R.R.; Green, R.; Padhee, S.; Howell, M.; Banerjee, J.; Mohapatra, S.S.; Mohapatra, S. Actinomycin D down-regulates SOX2 expression and induces death in breast cancer stem cells. Anticancer Res. 2017, 37, 1655–1663. [Google Scholar] [PubMed]
- Choong, M.L.; Yang, H.; Lee, M.A.; Lane, D.P. Specific activation of the p53 pathway by low dose actinomycin D: A new route to p53 based cyclotherapy. Cell Cycle 2009, 8, 2810–2818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saijets, S.; Ylipaasto, P.; Vaarala, O.; Hovi, T.; Roivainen, M. Enterovirus infection and activation of human umbilical vein endothelial cells. J. Med. Virol. 2003, 70, 430–439. [Google Scholar] [CrossRef]
- Rill, R.L.; Hecker, K.H. Sequence-specific actinomycin D binding to single-stranded DNA inhibits HIV reverse transcriptase and other polymerases. Biochemistry 1996, 35, 3525–3533. [Google Scholar] [CrossRef]
- Wang, D.; Wang, C.; Gui, P.; Liu, H.; Khalaf, S.M.H.; Elsayed, E.A.; Wadaan, M.A.M.; Hozzein, W.N.; Zhu, W. Identification, bioactivity and productivity of actinomycins from the marine-derived Streptomyces heliomycini. Front. Microbiol. 2017, 8, 1147. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Song, F.; Wang, Q.; Abdel-Mageed, W.M.; Guo, H.; Fu, C.; Hou, W.; Dai, H.; Liu, X.; Yang, N.; et al. A marine-derived Streptomyces sp. MS449 produces high yield of actinomycin X2 and actinomycin D with potent anti-tuberculosis activity. Appl. Microbiol. Biotechnol. 2012, 95, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Hammer, A.S.; Couto, C.G.; Ayl, R.D.; Shank, K.A. Treatment of tumor-bearing dogs with actinomycin D. J. Vet. Intern. Med. 1994, 8, 236–239. [Google Scholar] [CrossRef] [PubMed]
- Keller, U.; Lang, M.; Crnovcic, I.; Pfennig, F.; Schauwecker, F. The actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: A genetic hall of mirrors for synthesis of a molecule with mirror symmetry. J. Bacteriol. 2010, 192, 2583–2595. [Google Scholar] [CrossRef]
- Crnovčić, I.; Rückert, C.; Semsary, S.; Lang, M.; Kalinowski, J.; Keller, U. Genetic interrelations in the actinomycin biosynthetic gene clusters of Streptomyces antibioticus IMRU 3720 and Streptomyces chrysomallus ATCC11523, producers of actinomycin X and actinomycin C. Adv. Appl. Bioinform. Chem. 2017, 10, 29. [Google Scholar] [CrossRef]
- Wang, X.; Tabudravu, J.; Rateb, M.E.; Annand, K.J.; Qin, Z.; Jaspars, M.; Deng, Z.; Yu, Y.; Deng, H. Identification and characterization of the actinomycin G gene cluster in Streptomyces iakyrus. Mol. Biosyst. 2013, 9, 1286–1289. [Google Scholar] [CrossRef]
- Pfennig, F.; Schauwecker, F.; Keller, U. Molecular characterization of the genes of actinomycin synthetase I and of a 4-methyl-3-hydroxyanthranilic acid carrier protein involved in the assembly of the acylpeptide chain of actinomycin in Streptomyces. J. Biol. Chem. 1999, 274, 12508–12516. [Google Scholar] [CrossRef]
- Schauwecker, F.; Pfennig, F.; Grammel, N.; Keller, U. Construction and in vitro analysis of a new bi-modular polypeptide synthetase for synthesis of N-methylated acyl peptides. Chem. Biol. 2000, 7, 287–297. [Google Scholar] [CrossRef]
- Fawaz, F.; Jones, G.H. Actinomycin synthesis in Streptomyces antibioticus. Purification and properties of a 3-hydroxyanthranilate 4-methyltransferase. J. Biol. Chem. 1988, 4, 4602–4606. [Google Scholar]
- Smith, A.W.; Camara-Artigas, A.; Wang, M.; Allen, J.P.; Francisco, W.A. Structure of phenoxazinone synthase from Streptomyces antibioticus reveals a new type 2 copper center. Biochemistry 2006, 45, 4378–4387. [Google Scholar] [CrossRef]
- Madu, A.C.; Jones, G.H. Molecular cloning and in vitro expression of a silent phenoxazinone synthase gene from Streptomyces lividans. Gene 1989, 84, 287–294. [Google Scholar] [CrossRef]
- Golub, E.E.; Ward, M.A.; Nishimura, J.S. Biosynthesis of the actinomycin chromophore: Incorporation of 3-hydroxy-4-methylanthranilic acid into actinomycins by Streptomyces antibioticus. J. Bacteriol. 1969, 100, 977–984. [Google Scholar] [PubMed]
- Jones, G.H. Actinomycin production persists in a strain of Streptomyces antibioticus lacking phenoxazinone synthase. Antimicrob. Agents. Chemother. 2000, 44, 1322–1327. [Google Scholar] [CrossRef]
- Song, X.; Jiang, X.; Sun, J.; Zhang, C.; Zhang, Y.; Lu, C.; Ju, J. Antibacterial secondary metabolites produced by mangrove-derived actinomycete Stremptomeces costaricanus SCSIO ZS0073. Nat. Prod. Res. Dev. 2017, 29, 410–414. [Google Scholar]
- Jia, Y.; Xie, Y.; Li, Q.; Ma, J.; Huang, H.; Ju, J. Identification of the fungichromin biosynthetic pathway from Streptomyces costaricanus SCSIO ZS0073. Chin. J. Mar. Drug 2017, 36, 1–10. [Google Scholar]
- Baltz, R.H. Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J. Ind. Microbiol. Biotechnol. 2011, 38, 1747–1760. [Google Scholar] [CrossRef]
- Felnagle, E.A.; Barkei, J.J.; Park, H.; Podevels, A.M.; McMahon, M.D.; Drott, D.W.; Thomas, M.G. MbtH-like proteins as integral components of bacterial nonribosomal peptide synthetases. Biochemistry 2010, 49, 8815–8817. [Google Scholar] [CrossRef]
- Imker, H.J.; Krahn, D.; Clerc, J.; Kaiser, M.; Walsh, C.T. N-Acylation during glidobactin biosynthesis by the tridomain nonribosomal peptide synthetase module GlbF. Chem. Biol. 2010, 17, 1077–1083. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Genomics and the ancient origins of the daptomycin biosynthetic gene cluster. J. Antibiot. 2010, 63, 506–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quadri, L.E.; Sello, J.; Keating, T.A.; Weinreb, P.H.; Walsh, C.T. Identification of Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence conferring siderophore mycobactin. Chem. Biol. 1998, 5, 631–645. [Google Scholar] [CrossRef]
- Crnovčić, I.; Süssmuth, R.; Keller, U. Aromatic C-methyltransferases with antipodal stereoselectivity for structurally diverse phenolic amino acids catalyze the methylation step in the biosynthesis of the actinomycin chromophore. Biochemistry 2010, 49, 9698–9705. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [PubMed]
- Bassler, B.L.; Losick, R. Bacterially speaking. Cell 2006, 125, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.L.; Martínez-Bueno, M.; Molina-Henares, A.J.; Terán, W.; Watanabe, K.; Zhang, X.; Gallegos, M.T.; Brennan, R.; Tobes, R. The tetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005, 69, 326–356. [Google Scholar] [CrossRef]
- Nishida, H.; Ohnishi, Y.; Beppu, T.; Horinouchi, S. Evolution of γ-butyrolactone synthases and receptors in Streptomyces. Environ. Microbiol. 2007, 9, 1986–1994. [Google Scholar] [CrossRef] [PubMed]
- Gou, L.; Han, T.; Wang, X.; Ge, J.; Liu, W.; Hu, F.; Wang, Z. A novel tetR family transcriptional regulator, CalR3, negatively controls calcimycin biosynthesis in Streptomyces chartreusis NRRL 3882. Front. Microbiol. 2017, 8, 2371. [Google Scholar] [CrossRef]
- Mo, X.; Wang, Z.; Wang, B.; Ma, J.; Huang, H.; Tian, X.; Zhang, S.; Zhang, C.; Ju, J. Cloning and characterization of the biosynthetic gene cluster of the bacterial RNA polymerase inhibitor tirandamycin from marine-derived Streptomyces sp. SCSIO 1666. Biochem. Biophys. Res. Commun. 2011, 406, 341–347. [Google Scholar] [CrossRef]
- Peschke, U.; Schmidt, H.; Zhang, H.Z.; Pieperserg, W. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11. Mol. Microbiol. 1995, 16, 1137–1156. [Google Scholar] [CrossRef] [PubMed]
- Semsary, S.; Crnovčić, I.; Driller, R.; Vater, J.; Loll, B.; Keller, U. Ketonization of proline residues in the peptide chains of actinomycins by a 4-oxoproline synthase. ChemBioChem 2018, 19, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Waksman, S.A.; Woodruff, H.B. Bacteriostatic and bactericidal substances produced by a soil actinomyces. Exp. Biol. Med. 1940, 45, 609–614. [Google Scholar] [CrossRef]
- Gust, B.; Kieser, T.; Chater, K. PCR targeting system in Streptomyces coelicolor A3 (2). John Innes Centre 2002, 3, 1–39. [Google Scholar]




| Gene | Size a | Protein Homolog and Origin | ID/SM (%) | Origin (Protein ID) |
|---|---|---|---|---|
| orf(−3) | 492 | type VII secretion protein EccB | 96/97 | SCF88120 Streptomyces sp. LamerLS-31b |
| orf(−2) | 464 | type VII secretion integral membrane protein EccD | 99/99 | SCF88128 Streptomyces sp. LamerLS-31b |
| orf(−1) | 1289 | Collagen triple helix repeat-containing protein | 53/75 | Clostridium uliginosum |
| acnW | 103 | Hypothetical protein | 99/100 | AB905443.1 Streptomyces rochei 7434AN4 |
| acnA | 121 | Protein of unknown function | 93/96 | SCF88143 Streptomyces sp. LamerLS-31b |
| acnB | 415 | Anti-anti-sigma regulatory factor | 96/97 | SCF88151 Streptomyces sp. LamerLS-31b |
| acnU1 | 121 | hypothetical protein GA0115258_11557 | 99/100 | SCF88158 Streptomyces sp. LamerLS-31b |
| acnU2 | 124 | hypothetical protein GA0115258_11558 | 95/96 | SCF88164 Streptomyces sp. LamerLS-31b |
| acnC | 392 | acyl-CoA dehydrogenase | 91/95 | AKJ14982 Streptomyces incarnatus |
| acnU3 | 207 | hypothetical protein | 80/91 | WP_030987180 Streptomyces |
| acnU4 | 187 | hypothetical protein | 82/88 | WP_030592054 Streptomyces anulatus |
| acnD | 66 | protein mbtH | 91/95 | OOQ48080 Streptomyces antibioticus |
| acnE | 78 | 4-MHA carrier protein | 73/85 | ADG27356 Streptomyces anulatus |
| acnN1 | 467 | Adenylation domain protein | 51/65 | SCD96508 Streptomyces sp. DvalAA-43 |
| acnN2 | 2589 | non-ribosomal peptide synthase | 73/81 | WP_057667184 Streptomyces anulatus |
| acnN3 | 4249 | non-ribosomal peptide synthetase | 78/85 | WP_064726364 Streptomyces parvulus |
| acnF | 211 | hypothetical protein | 84/88 | OOQ48467 Streptomyces antibioticus |
| acnG | 326 | arylformamidase | 79/85 | SCF58504 Streptomyces sp. Cmuel-A718b |
| acnH | 284 | tryptophan 2, 3-dioxygenase | 84/88 | OOQ48467 Streptomyces antibioticus |
| acnL | 420 | kynureninase | 86/90 | OOQ48077 Streptomyces antibioticus |
| acnM | 346 | methyltransferase | 88/91 | WP_030594248 Streptomyces anulatus |
| acnP | 386 | cytochrome P450 | 86/91 | WP_030594247 Streptomyces anulatus |
| acnO | 224 | LmbU-like protein | 73/82 | ADG27350 Streptomyces anulatus |
| acnR | 282 | TetR family transcriptional regulator | 78/86 | OOQ48074 Streptomyces antibioticus |
| acnQ | 294 | siderophore-interacting protein | 78/87 | WP_030594239 Streptomyces anulatus |
| acnT1 | 346 | ABC transporter | 87/92 | OOQ48072 Streptomyces antibioticus |
| acnT2 | 255 | multidrug ABC transporter permease | 92/95 | OOQ48071 Streptomyces antibioticus |
| acnT3 | 753 | UvrABC system protein A | 60/77 | AMV28079 Gemmata sp. SH-PL17 |
| orf(+1) | 191 | GCN5-related N-acetyltransferase | 81/87 | UN35851 Streptomyces venezuelae |
| orf(+2) | 109 | hypothetical protein | 74/83 | WP_052876109 Streptomyces sp. NRRL F-4335 |
| orf(+3) | 1130 | Two-component hybrid sensor and regulator | 55/68 | SBO98167 Nonomuraea sp. ATCC 39727 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Jia, Y.; Xie, Y.; Zhang, C.; Ma, J.; Sun, C.; Ju, J. Identification of the Actinomycin D Biosynthetic Pathway from Marine-Derived Streptomyces costaricanus SCSIO ZS0073. Mar. Drugs 2019, 17, 240. https://doi.org/10.3390/md17040240
Liu M, Jia Y, Xie Y, Zhang C, Ma J, Sun C, Ju J. Identification of the Actinomycin D Biosynthetic Pathway from Marine-Derived Streptomyces costaricanus SCSIO ZS0073. Marine Drugs. 2019; 17(4):240. https://doi.org/10.3390/md17040240
Chicago/Turabian StyleLiu, Mengchan, Yanxi Jia, Yunchang Xie, Chunyan Zhang, Junying Ma, Changli Sun, and Jianhua Ju. 2019. "Identification of the Actinomycin D Biosynthetic Pathway from Marine-Derived Streptomyces costaricanus SCSIO ZS0073" Marine Drugs 17, no. 4: 240. https://doi.org/10.3390/md17040240
APA StyleLiu, M., Jia, Y., Xie, Y., Zhang, C., Ma, J., Sun, C., & Ju, J. (2019). Identification of the Actinomycin D Biosynthetic Pathway from Marine-Derived Streptomyces costaricanus SCSIO ZS0073. Marine Drugs, 17(4), 240. https://doi.org/10.3390/md17040240






