Identification of Antimicrobial Peptides from the Microalgae Tetraselmis suecica (Kylin) Butcher and Bactericidal Activity Improvement
Abstract
:1. Introduction
2. Results
2.1. Characterization and Antimicrobial Activity of Acid Extracts from Tetraselmis suecica
2.2. Peptide De Novo Sequence and Antibacterial Activity
2.3. Structural Characterization and Cytotoxicity Analysis of Antimicrobial Peptides of T. suecica
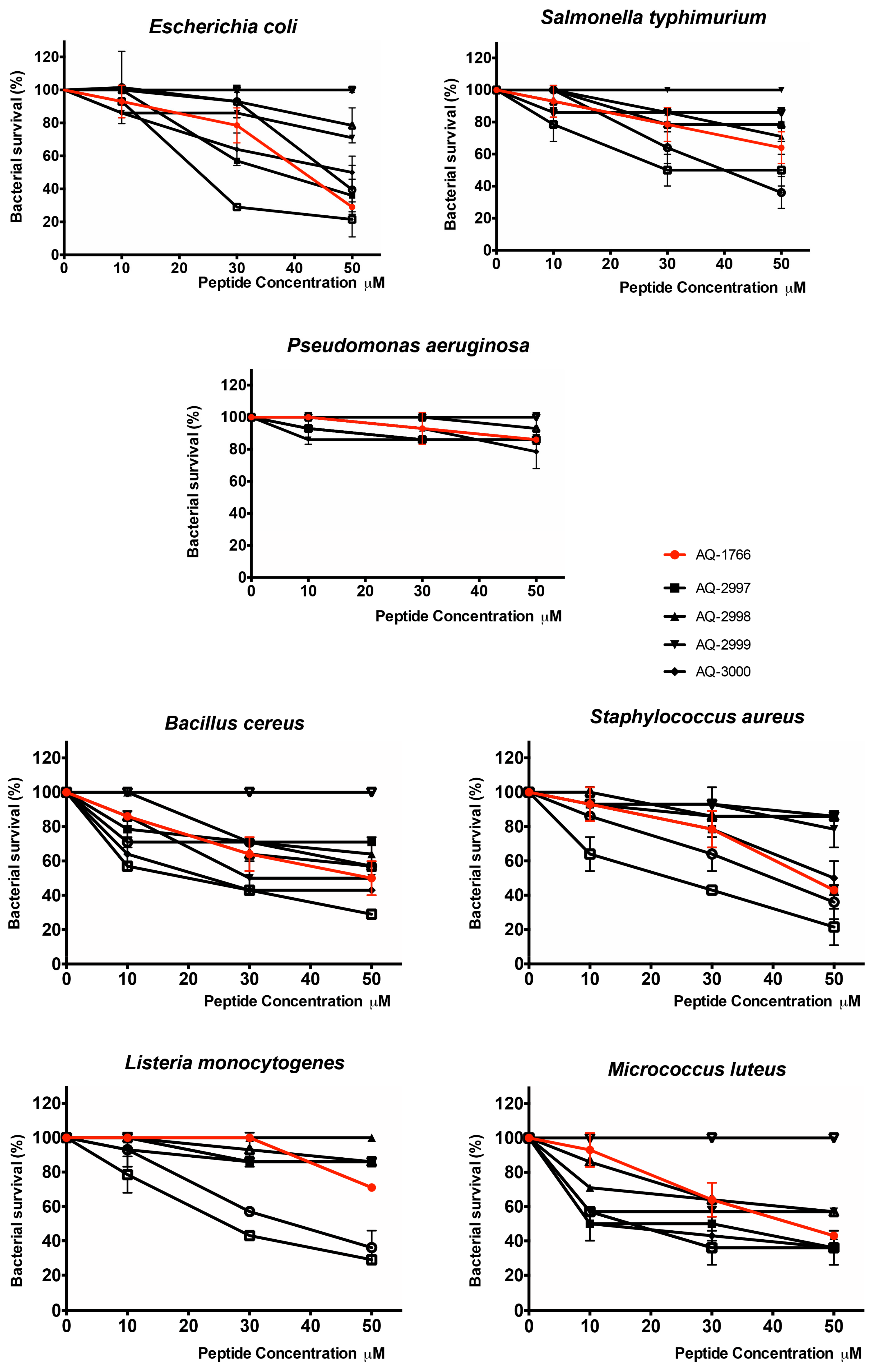
2.4. Characterization and Activity of Analog Peptides
2.4.1. Antibacterial Activity of AQ-1766 Alanine Scan Analogs
2.4.2. Antibacterial Activity of AQ-1766 Lysine Scan Analogs
2.4.3. Secondary Structure Analysis of AQ-1766 Alanine and Lysine Scan Analogs
2.4.4. Cytotoxicity Analysis of AQ-1766 Alanine and Lysine Scan Analogs
3. Discussion
4. Materials and Methods
4.1. Tetraselmis Suecica Culture
4.2. Production of Acid Extracts
4.3. Enrichment and Characterization of T. suecica Extracts
4.4. Characterization of the Enriched Peptide Fractions by Gel Electrophoresis
4.5. Antibacterial Assays
4.6. Mass Spectrometry
4.7. LC-ESI-MS/MS Mass Spectrometry
4.8. Peptide Synthesis and Purification
4.9. Secondary Structure Analysis
4.10. Peptide Modifications by Alanine Scan or Lysine Substitution
4.11. Cytotoxic Activity of Synthetic Peptides
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Abu-Ghannam, N.; Rajauria, G. Antimicrobial activity of compounds isolated from algae. In Functional Ingredients from Algae for Foods and Nutraceuticals; Dominguez, H., Ed.; Woodhead Publishing Limited: Cambridge, UK, 2013; pp. 287–306. ISBN 9780857095121. [Google Scholar]
- Guedes, A.C.; Amaro, H.M.; Malcata, F.X. Microalgae as sources of high added-value compounds—A brief review of recent work. Biotechnol. Prog. 2011, 27, 597–613. [Google Scholar] [CrossRef] [PubMed]
- Mimouni, V.; Ulmann, L.; Pasquet, V.; Mathieu, M.; Picot, L.; Bougaran, G.; Cadoret, J.-P.; Morant-Manceau, A.; Schoefs, B. The Potential of Microalgae for the Production of Bioactive Molecules of Pharmaceutical Interest. Curr. Pharm. Biotechnol. 2012, 13, 2733–2750. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chen, L.; Zhang, W. Chemicals to enhance microalgal growth and accumulation of high-value bioproducts. Front. Microbiol. 2015, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cabello, F.C. Antibiotics and aquaculture in Chile: Implications for human and animal health. Rev. Med. Chil. 2004, 132, 1001–1006. [Google Scholar] [PubMed]
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Guilhelmelli, F.; Vilela, N.; Albuquerque, P.; Derengowski, L.d.S.; Silva-Pereira, I.; Kyaw, C.M. Antibiotic development challenges: The various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Smith, V.J.; Desbois, A.P.; Dyrynda, E.A. Conventional and Unconventional Antimicrobials from Fish, Marine Invertebrates and Micro-algae. Mar. Drugs 2010, 8, 1213–1262. [Google Scholar] [CrossRef] [Green Version]
- Falaise, C.; François, C.; Travers, M.-A.; Morga, B.; Haure, J.; Tremblay, R.; Turcotte, F.; Pasetto, P.; Gastineau, R.; Hardivillier, Y.; et al. Antimicrobial Compounds from Eukaryotic Microalgae against Human Pathogens and Diseases in Aquaculture. Mar. Drugs 2016, 14, 159. [Google Scholar] [CrossRef]
- Pane, G.; Cacciola, G.; Giacco, E.; Mariottini, G.; Coppo, E. Assessment of the Antimicrobial Activity of Algae Extracts on Bacteria Responsible of External Otitis. Mar. Drugs 2015, 13, 6440–6452. [Google Scholar] [CrossRef]
- Kim, S.-K.; Wijesekara, I. Development and biological activities of marine-derived bioactive peptides: A review. J. Funct. Foods 2010, 2, 1–9. [Google Scholar] [CrossRef]
- Samarakoon, K.; Jeon, Y.-J. Bio-functionalities of proteins derived from marine algae—A review. Food Res. Int. 2012, 48, 948–960. [Google Scholar] [CrossRef]
- Ruiz-Ruiz, F.; Mancera-Andrade, E.I.; Iqbal, H.M. Marine-Derived Bioactive Peptides for Biomedical Sectors: A Review. Protein Pept. Lett. 2017, 24, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kizhakkedathu, J.; Straus, S. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Montone, C.M.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Piovesana, S.; Zenezini Chiozzi, R.; Laganà, A. Peptidomic strategy for purification and identification of potential ACE-inhibitory and antioxidant peptides in Tetradesmus obliquus microalgae. Anal. Bioanal. Chem. 2018, 410, 3573–3586. [Google Scholar] [CrossRef] [PubMed]
- Giordano, D.; Costantini, M.; Coppola, D.; Lauritano, C.; Núñez Pons, L.; Ruocco, N.; di Prisco, G.; Ianora, A.; Verde, C. Biotechnological Applications of Bioactive Peptides From Marine Sources. Adv. Microb. Physiol. 2018, 73, 171–220. [Google Scholar]
- Duff, D.; Bruce, D.; Antia, N. The Antibacterial Activity of Marine Planktonic Algae. Can. J. Microbiol. 1966, 12, 877–884. [Google Scholar] [CrossRef]
- Austin, B.; Day, J.G. Inhibition of prawn pathogenic Vibrio spp. by a commercial spray-dried preparation of Tetraselmis suecica. Aquaculture 1990, 90, 389–392. [Google Scholar] [CrossRef]
- Austin, B.; Baudet, E.; Stobie, M. Inhibition of bacterial fish pathogens by Tetraselmis suecica. J. Fish Dis. 1992, 15, 55–61. [Google Scholar] [CrossRef]
- Morrison, K.L.; Weiss, G.A. Combinatorial alanine-scanning. Curr. Opin. Chem. Biol. 2001, 5, 302–307. [Google Scholar] [CrossRef]
- Weiss, G.A.; Watanabe, C.K.; Zhong, A.; Goddard, A.; Sidhu, S.S. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc. Natl. Acad. Sci. USA 2000, 97, 8950–8954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, H.; Feix, J.B. Lysine-Enriched Cecropin-Mellitin Antimicrobial Peptides with Enhanced Selectivity. Antimicrob. Agents Chemother. 2008, 52, 4463–4465. [Google Scholar] [CrossRef] [PubMed]
- Almaaytah, A.; Tarazi, S.; Abu-Alhaijaa, A.; Altall, Y.; Alshar’i, N.; Bodoor, K.; Al-Balas, Q. Enhanced Antimicrobial Activity of AamAP1-Lysine, a Novel Synthetic Peptide Analog Derived from the Scorpion Venom Peptide AamAP1. Pharmaceuticals 2014, 7, 502–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitta, G.; Hubert, F.; Noel, T.; Roch, P. Myticin, a novel cysteine-rich antimicrobial peptide isolated from haemocytes and plasma of the mussel Mytilus galloprovincialis. Eur. J. Biochem. 1999, 265, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, C.A.; Barriga, A.; Albericio, F.; Romero, M.S.; Guzmán, F. Identification of Peptides in Flowers of Sambucus nigra with Antimicrobial Activity against Aquaculture Pathogen. Molecules 2018, 23. [Google Scholar] [CrossRef] [PubMed]
- Intiquilla, A.; Jiménez-Aliaga, K.; Guzmán, F.; Alvarez, C.A.; Zavaleta, A.I.; Izaguirre, V.; Hernández-Ledesma, B. Novel antioxidant peptides obtained by alcalase hydrolysis of Erythrina edulis (pajuro) protein. J. Sci. Food Agric. 2019, 99, 2420–2427. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, P.; Wacyk, J.; Morales-Lange, B.; Rojas, V.; Guzmán, F.; Dixon, B.; Mercado, L. Immunomodulatory effect of cathelicidins in response to a β-glucan in intestinal epithelial cells from rainbow trout. Dev. Comp. Immunol. 2015, 51, 160–169. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Riss, T.; Moravec, R.; Niles, A.; Duellman, S.; Benink, H.; Worzella, T.; Minor, L. Cell Viability Assays. In Assay Guidance Manual [Internet]; Sittampalam, G.S., Grossman, A., Brimacombe, K., Arkin, M., Auld, D., Austin, C., Baell, J., Bejcek, B., Caaveiro, J.J.M., Chung, T.D.Y., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. [Google Scholar]
- Rozek, T.; Wegener, K.L.; Bowie, J.H.; Olver, I.N.; Carver, J.A.; Wallace, J.C.; Tyler, M.J. The antibiotic and anticancer active aurein peptides from the Australian Bell Frogs Litoria aurea and Litoria raniformis. Eur. J. Biochem. 2000, 267, 5330–5341. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Toniolo, C.; Bonora, G.M. Structural Aspects of Small Peptides. A Circular Dichroism Study of Monodisperse Protected Homo-Oligomers Derived from L-Alanine. Die Makromol. Chem./Macromol. Chem. 1975, 176, 2547–2558. [Google Scholar] [CrossRef]
- Mahalakshmi, R.; Sengupta, A.; Raghothama, S.; Shamala, N.; Balaram, P. Tryptophan-containing peptide helices: Interactions involving the indole side chain. J. Pept. Res. 2005, 66, 277–296. [Google Scholar] [CrossRef] [PubMed]
- Sedighi, M.; Jalili, H.; Darvish, M.; Sadeghi, S.; Ranaei-Siadat, S.-O. Enzymatic hydrolysis of microalgae proteins using serine proteases: A study to characterize kinetic parameters. Food Chem. 2019, 284, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Tejano, L.A.; Peralta, J.P.; Yap, E.E.S.; Chang, Y. Bioactivities of enzymatic protein hydrolysates derived from Chlorella sorokiniana. Food Sci. Nutr. 2019. [Google Scholar] [CrossRef] [PubMed]
- Shannon, E.; Abu-Ghannam, N. Antibacterial Derivatives of Marine Algae: An Overview of Pharmacological Mechanisms and Applications. Mar. Drugs 2016, 14, 81. [Google Scholar] [CrossRef] [PubMed]
- Suetsuna, K.; Chen, J.R. Identification of antihypertensive peptides from peptic digest of two microalgae, Chlorella vulgaris and Spirulina platensis. Mar. Biotechnol. 2001, 3, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Sheih, I.-C.; Fang, T.J.; Wu, T.-K. Isolation and characterisation of a novel angiotensin I-converting enzyme (ACE) inhibitory peptide from the algae protein waste. Food Chem. 2009, 115, 279–284. [Google Scholar] [CrossRef]
- Kang, K.H.; Ryu, B.M.; Kim, S.K.; Qian, Z.J. Characterization of growth and protein contents from microalgae Navicula incerta with the investigation of antioxidant activity of enzymatic hydrolysates. Food Sci. Biotechnol. 2011, 20, 183–191. [Google Scholar] [CrossRef]
- Sheih, I.-C.; Fang, T.J.; Wu, T.-K.; Lin, P.-H. Anticancer and Antioxidant Activities of the Peptide Fraction from Algae Protein Waste. J. Agric. Food Chem. 2010, 58, 1202–1207. [Google Scholar] [CrossRef]
- Ejike, C.E.C.C.; Collins, S.A.; Balasuriya, N.; Swanson, A.K.; Mason, B.; Udenigwe, C.C. Prospects of microalgae proteins in producing peptide-based functional foods for promoting cardiovascular health. Trends Food Sci. Technol. 2017, 59, 30–36. [Google Scholar] [CrossRef]
- Carvajal-Rondanelli, P.; Aróstica, M.; Álvarez, C.A.; Ojeda, C.; Albericio, F.; Aguilar, L.F.; Marshall, S.H.; Guzmán, F. Understanding the antimicrobial properties/activity of an 11-residue Lys homopeptide by alanine and proline scan. Amino Acids 2018, 50, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Mahalakshmi, R.; Shanmugam, G.; Polavarapu, P.L.; Balaram, P. Circular Dichroism of Designed Peptide Helices and β-Hairpins: Analysis of Trp- and Tyr-Rich Peptides. ChemBioChem 2005, 6, 2152–2158. [Google Scholar] [CrossRef] [PubMed]
- Won, H.-S.; Seo, M.-D.; Jung, S.-J.; Lee, S.-J.; Kang, S.-J.; Son, W.-S.; Kim, H.-J.; Park, T.-K.; Park, S.-J.; Lee, B.-J. Structural Determinants for the Membrane Interaction of Novel Bioactive Undecapeptides Derived from Gaegurin 5. J. Med. Chem. 2006, 49, 4886–4895. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Kim, S.-J.; Lee, Y.-S.; Song, M.-D.; Kim, I.-H.; Won, H.-S. De novo generation of short antimicrobial peptides with simple amino acid composition. Regul. Pept. 2011, 166, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Hoernke, M.; Schwieger, C.; Kerth, A.; Blume, A. Binding of cationic pentapeptides with modified side chain lengths to negatively charged lipid membranes: Complex interplay of electrostatic and hydrophobic interactions. Biochim. Biophys. Acta 2012, 1818, 1663–1672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef]
- Da SilvaVaz, B.; Moreira, J.B.; de Morais, M.G.; Costa, J.A.V. Microalgae as a new source of bioactive compounds in food supplements. Curr. Opin. Food Sci. 2016, 7, 73–77. [Google Scholar]
- Cabello, F.C. Antibióticos y acuicultura en Chile: Consecuencias para la salud humana y animal. Rev. Med. Chile 2004, 132, 1001–1006. [Google Scholar] [CrossRef]
- Wang, S.; Zeng, X.; Yang, Q.; Qiao, S. Antimicrobial Peptides as Potential Alternatives to Antibiotics in Food Animal Industry. Int. J. Mol. Sci. 2016, 17, 603. [Google Scholar] [CrossRef]
- Guillard, R.; Ryther, J. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef]
- Paniagua Michel, J.; Buckle Ramirez, L.F.; Granados, M.C.; Loya Salinas, D.H. Manual de Metodologías y Alternativas Para el Cultivo de Microalgas; CICESE: Ensenada, BC, Mexico, 1989. [Google Scholar]
- Schägger, H. Tricine–SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Mastroianni, J.R.; Ouellette, A.J. α-Defensins in Enteric Innate Immunity. J. Biol. Chem. 2009, 284, 27848–27856. [Google Scholar] [CrossRef] [PubMed]
- Segura, C.; Guzmán, F.; Salazar, L.M.; Patarroyo, M.E.; Orduz, S.; Lemeshko, V. BTM-P1 polycationic peptide biological activity and 3D-dimensional structure. Biochem. Biophys. Res. Commun. 2007, 353, 908–914. [Google Scholar] [CrossRef] [PubMed]
- Sunarintyas, S.; Siswomihardjo, W.; Tontowi, A.E. Cytotoxicity of Cricula triphenestrata Cocoon Extract on Human Fibroblasts. Int. J. Biomater. 2012, 2012, 1–5. [Google Scholar] [CrossRef] [PubMed]





| Bacteria | Bacterial Cell Survival (%) a | ||||
|---|---|---|---|---|---|
| 40% ACN Peptide Concentration | BTM P1 b (C+) 0.06 μg/μL | ||||
| 0.1 μg/μL | 0.3 μg/μL | 0.5 μg/μL | |||
| Gram− | Escherichia coli ML35 | 56 | 23 | 4 | 0 |
| Salmonella typhimurium ATCC 14028 | 93 | 82 | 78 | 7 | |
| Pseudomonas aeruginosa ATCC 27853 | 100 | 93 | 93 | 11 | |
| Gram+ | Bacillus cereus ISP B7/13 | 52 | 0 | 0 | 0 |
| Staphylococcus aureus ATCC 25933 | 89 | 48 | 19 | 4 | |
| Listeria monocytogenes ATCC 19115 | 100 | 82 | 93 | 11 | |
| Micrococcus luteus ATCC 9341 | 100 | 96 | 96 | 7 | |
| Alanine Analogs | Lysine Analogs | ||
|---|---|---|---|
| Peptide | Sequence | Peptide | Sequence |
| AQ-1766 | LWFYTMWH | AQ-1766 | LWFYTMWH |
| AQ-2997 | AWFYTMWH | AQ-3369 | KWFYTMWH |
| AQ-2998 | LAFYTMWH | ||
| AQ-2999 | LWAYTMWH | ||
| AQ-3000 | LWFATMWH | AQ-3370 | LWFKTMWH |
| AQ-3001 | LWFYAMWH | AQ-3371 | LWFYKMWH |
| AQ-3002 | LWFYTAWH | AQ-3372 | LWFYTKWH |
| AQ-3003 | LWFYTMAH | ||
| AQ-3004 | LWFYTMWA | ||
| Peptide | AQ-1766 | AQ-2997 | AQ-2998 | AQ-2999 | AQ-3000 | AQ-3001 | AQ-3002 | AQ-3003 | AQ-3004 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | ||||||||||
| Gram− | E. coli ML35 | 41 | 36 | NA | >50 | 50 | 46 | 23 *** | >50 | NA |
| S. typhimurium ATCC 14028 | >50 | >50 | >50 | NA | >50 | 40 | 30 | >50 | >50 | |
| P. aeruginosa ATCC 27853 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | |
| Gram+ | B. cereus ISP B7/13 | 50 | >50 | >50 | 30 ** | 23 *** | >50 | 20 **** | >50 | NA |
| S. aureus ATCC 25933 | 46 | 46 | >50 | >50 | 50 | 43 | 23 *** | >50 | >50 | |
| L. monocytogenes ATCC 19115 | >50 | >50 | >50 | >50 | >50 | 36 | 26 | >50 | >50 | |
| M. luteus ATCC 9341 | 44 | 10 | 44 | >50 | 10 * | 17 * | 17 * | >50 | NA |
| Peptide | AQ-1766 | AQ-3369 | AQ-3370 | AQ-3371 | AQ-3372 | |
|---|---|---|---|---|---|---|
| Bacteria | ||||||
| Gram− | E. coli ML35 | 41 | 8 **** | 20 *** | 23 *** | 42 |
| S. typhimurium ATCC 14028 | >50 | 30 | 40 | >50 | >50 | |
| P. aeruginosa ATCC 27853 | >50 | >50 | >50 | >50 | >50 | |
| Gram+ | B. cereus ISP B7/13 | 50 | 10 **** | 20 **** | >50 | 43 |
| S. aureus ATCC 25933 | 46 | 19 *** | 25 ** | 30 * | 44 | |
| L. monocytogenes ATCC 19115 | >50 | 25 | 46 | 30 | 50 | |
| M. luteus ATCC 9341 | 44 | 30 | 44 | 30 | >50 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guzmán, F.; Wong, G.; Román, T.; Cárdenas, C.; Alvárez, C.; Schmitt, P.; Albericio, F.; Rojas, V. Identification of Antimicrobial Peptides from the Microalgae Tetraselmis suecica (Kylin) Butcher and Bactericidal Activity Improvement. Mar. Drugs 2019, 17, 453. https://doi.org/10.3390/md17080453
Guzmán F, Wong G, Román T, Cárdenas C, Alvárez C, Schmitt P, Albericio F, Rojas V. Identification of Antimicrobial Peptides from the Microalgae Tetraselmis suecica (Kylin) Butcher and Bactericidal Activity Improvement. Marine Drugs. 2019; 17(8):453. https://doi.org/10.3390/md17080453
Chicago/Turabian StyleGuzmán, Fanny, Genezareth Wong, Tanya Román, Constanza Cárdenas, Claudio Alvárez, Paulina Schmitt, Fernando Albericio, and Verónica Rojas. 2019. "Identification of Antimicrobial Peptides from the Microalgae Tetraselmis suecica (Kylin) Butcher and Bactericidal Activity Improvement" Marine Drugs 17, no. 8: 453. https://doi.org/10.3390/md17080453
APA StyleGuzmán, F., Wong, G., Román, T., Cárdenas, C., Alvárez, C., Schmitt, P., Albericio, F., & Rojas, V. (2019). Identification of Antimicrobial Peptides from the Microalgae Tetraselmis suecica (Kylin) Butcher and Bactericidal Activity Improvement. Marine Drugs, 17(8), 453. https://doi.org/10.3390/md17080453