Scopularides Revisited: Molecular Networking Guided Exploration of Lipodepsipeptides in Australian Marine Fish Gastrointestinal Tract-Derived Fungi
Abstract
:1. Introduction
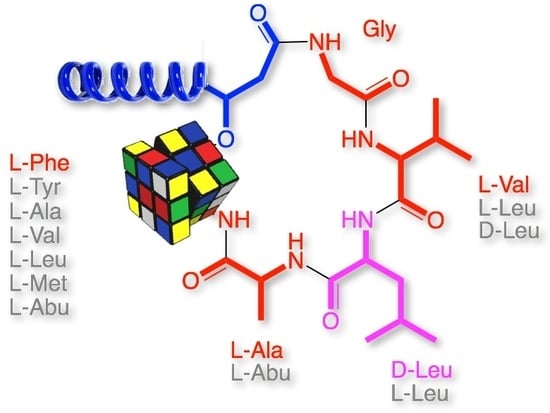
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Strain Isolation and Taxonomy
3.3. Scale up Cultivation of Scopulariopsis sp. CMB-F458
3.4. Scale-Up Cultivation of Beauveria sp. CMB-F585
3.5. Scale-Up Cultivation of Scopulariopsis sp. CMB-F115
3.6. Metabolite Charcterization
- Scopularide A (1); white powder; [α]D22.8 − 34.9 (c 0.25, MeOH); NMR (600 MHz, MeOH-d4) see Table S1, Figures S6 and S7; NMR (600 MHz, DMSO-d6) see Table S2, Figures S8 and S9; ESI(+)MS m/z 672 [M + H]+; HRESI(+)MS m/z 694.4149 [M + Na]+ (calcd. for C36H57N5O7Na, 694.4150); GNPS library CCMSLIB00005436489.
- Scopularide B (2); white powder; [α]D22.2 − 19.8 (c 0.05, MeOH); NMR (600 MHz, MeOH-d4) see Table S3, Figures S10 and S11; NMR (600 MHz, DMSO-d6) see Table S4, Figures S12 and S13; ESI(+)MS m/z 644 [M + H]+; HRESI(+)MS m/z 666.3842 [M + Na]+ (calcd. for C34H53N5O7Na, 666.3837); GNPS library CCMSLIB00005436490.
- Scopularide C (3); white powder; [α]D22.2 − 32.0 (c 0.13, MeOH); NMR (600 MHz, DMSO-d6) see Table 1 and Table S5, Figures S14–S18; ESI(+)MS m/z 714 [M + H]+; HRESI(+)MS m/z 736.4638 [M + Na]+ (calcd. for C39H63N5O7Na, 736.4620); GNPS library CCMSLIB00005436483.
- Scopularide D (4); white powder; [α]D21.9 − 40.9 (c 0.13, MeOH); NMR (600 MHz, DMSO-d6) see Table 1 and Table S6, Figures S19–S23; ESI(+)MS m/z 666 [M + H]+; HRESI(+)MS m/z 688.4620 [M + Na]+ (calcd. for C35H63N5O7Na, 688.4620); GNPS library CCMSLIB00005436484.
- Scopularide E (5); white powder; [α]D21.6 − 19.2 (c 0.13, MeOH); NMR (600 MHz, DMSO-d6) see Table 1 and Table S7, Figures S24–S28; ESI(+)MS m/z 638 [M + H]+; HRESI(+)MS m/z 660.4313 [M + Na]+ (calcd. for C33H59N5O7Na, 660.4307); GNPS library CCMSLIB00005436485.
- Scopularide F (6); white powder; [α]D24.2 − 17.5 (c 0.1, MeOH); NMR (600 MHz, DMSO-d6) see Table 2 and Table S8, Figures S29–S33; ESI(+)MS m/z 728 [M + H]+; HRESI(+)MS m/z 750.4787 [M + Na]+ (calcd. for C40H65N5O7Na, 750.4776); GNPS library CCMSLIB00005436486.
- Scopularide G (7); white powder; ESI(+)MS m/z 680 [M + H]+; HRESI(+)MS m/z 702.4789 [M + Na]+ (calcd. for C36H65N5O7Na, 702.4776) GNPS library CCMSLIB00005436487.
- Scopularide H (8); white powder; [α]D22.2 − 31.0 (c 0.15, MeOH); NMR (600 MHz, DMSO-d6) see Table 2 and Table S9, Figures S34–S38; ESI(+)MS m/z 700 [M + H]+; HRESI(+)MS m/z 722.4464 [M + Na]+ (calcd. for C38H61N5O7Na, 722.4463); GNPS library CCMSLIB00005436488.
3.7. C3 Marfey’s Analyses
3.8. Global Natural Product Social (GNPS) Molecular Networking
- Fish microbial library molecular networkingMASSIVE: MSV000084190 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=479a543e9b45408bbe93414769058784).
- CMB-F585 molecular networkingMASSIVE: MSV000084191 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=4dee2595568c4296a1c4393de4758b24).
- GNPS Molecular Networking job: (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=ae56648aefa54f789e04cfc314cb59fc).
- CMB-F115 molecular networkingMASSIVE: MSV000084192 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=74da14126ec24f2ca62ba443a05af7d2).
- GNPS Molecular Networking job: (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=e90bdf9596ff4bbfb9e4eb200435a2b1).
- CMB-F458 molecular networkingMASSIVE: MSV000084193 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=4b0ec7ee53c34a299fe1828588ead84d).
- GNPS Molecular Networking: (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=3c2af674f0a248c39149993d91a8149e)
3.9. X-Ray Analysis of Scopularides C (3) and H (8)
3.10. Antibacterial Assay
3.11. Antifungal Assay
3.12. Cytotoxicity Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mohamed, O.G.; Khalil, Z.G.; Capon, R.J. Prolinimines: N-Amino-L-Pro-methyl ester (hydrazine) Schiff bases from a fish gastrointestinal tract-derived fungus, Trichoderma sp. CMB-F563. Org. Lett. 2018, 20, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Lang, G.; Kajahn, I.; Schmaljohann, R.; Imhoff, J.F. Scopularides A and B, cyclodepsipeptides from a marine sponge-derived fungus, Scopulariopsis brevicaulis. J. Nat. Prod. 2008, 71, 1052–1054. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijayasarathy, S.; Prasad, P.; Fremlin, L.J.; Ratnayake, R.; Salim, A.A.; Khalil, Z.; Capon, R.J. C3 and 2D C3 Marfey’s methods for amino acid analysis in natural products. J. Nat. Prod. 2016, 79, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.-J.; Yuan, W.; Liao, X.-J.; Han, B.-N.; Wang, S.-P.; Li, Z.-Y.; Xu, S.-H.; Zhang, W.; Lin, H.-W. Oryzamides A–E, cyclodepsipeptides from the sponge-derived fungus Nigrospora oryzae PF18. J. Nat. Prod. 2016, 79, 2045–2052. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.-M.; Li, Y.-Y.; Shi, Y.-W.; Fang, Y.W.; Chao, R.; Gu, Y.-C.; Wang, C.-Y.; Shao, C.-L. Integrating molecular networking and 1H NMR to target the isolation of chrysogeamides from a library of marine-derived Penicillium fungi. J. Org. Chem. 2019, 84, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.-C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J. Nat. Prod. 2007, 70, 515–520. [Google Scholar] [CrossRef]
- Chiang, Y.-M.; Szewczyk, E.; Nayak, T.; Davidson, A.D.; Sanchez, J.F.; Lo, H.-C.; Ho, W.-Y.; Simityan, H.; Kuo, E.; Praseuth, A.; et al. Molecular genetic mining of the Aspergillus secondary metabolome: Discovery of the emericellamide biosynthetic pathway. Chem. Biol. 2008, 15, 527–532. [Google Scholar] [CrossRef]
- Wu, H.M.; Lin, L.P.; Xu, Q.L.; Han, W.B.; Zhang, S.; Liu, Z.W.; Mei, Y.N.; Yao, Z.-J.; Tan, R.X. Nodupetide, a potent insecticide and antimicrobial from Nodulisporium sp. associated with Riptortus pedestris. Tetrahedron Letts. 2017, 58, 663–665. [Google Scholar] [CrossRef]
- Vining, L.C.; Taber, W.A. Isariin, a new depsipeptide from Isaria cretacea. Can. J. Chem. 1962, 40, 1579–1584. [Google Scholar] [CrossRef]
- Ióca, L.P.; Nicacio, K.J.; Berlinck, R.G.S.; Ióca, L.P.; Nicacio, K.J.; Berlinck, R.G.S. Natural Products from marine invertebrates and microorganisms in Brazil between 2004 and 2017: Still the challenges, more rewards. J. Braz. Chem. Soc. 2018, 29, 998–1031. [Google Scholar] [CrossRef]
- Lira, S.P.; Vita-marques, A.M.; Seleghim, M.H.R.; Bugni, T.S.; Labarbera, D.V.; Sette, L.D.; Sponchiado, S.R.P.; Ireland, C.M.; Berlinck, R.G.S. New destruxins from the marine-derived fungus Beauveria felina. J. Antibiot. 2006, 59, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Baute, R.; Deffieux, G.; Merlet, D.; Baute, M.A.; Neveu, A. New insecticidal cyclodepsipeptides from the fungus Isaria felina: I. Production, isolation and insecticidal properties of isariins B, C and D. J. Antibiot. 1981, 34, 1261–1265. [Google Scholar] [CrossRef] [PubMed]
- Deffieux, G.; Merlet, D.; Baute, R.; Bourgeois, G.; Baute, M.A.; Neveu, A. New insecticidal cyclodepsipeptides from the fungus Isaria felina II. Structure elucidation of isariins B, C and D. J. Antibiot. 1981, 34, 1266–1270. [Google Scholar] [CrossRef] [PubMed]
- Sabareesh, V.; Ranganayaki, R.S.; Raghothama, S.; Bopanna, M.P.; Balaram, H.; Srinivasan, M.C.; Balaram, P. Identification and characterization of a library of microheterogeneous cyclohexadepsipeptides from the fungus Isaria. J. Nat. Prod. 2007, 70, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Langenfeld, A.; Blond, A.; Gueye, S.; Herson, P.; Nay, B.; Dupont, J.; Prado, S. Insecticidal cyclodepsipeptides from Beauveria felina. J. Nat. Prod. 2011, 74, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Du, F.-Y.; Li, X.-M.; Zhang, P.; Li, C.-S.; Wang, B.-G. Cyclodepsipeptides and other O-containing heterocyclic metabolites from Beauveria felina EN-135, a marine-derived entomopathogenic fungus. Mar. Drugs 2014, 12, 2816–2826. [Google Scholar] [CrossRef] [PubMed]
- Asolkar, R.N.; Freel, K.C.; Jensen, P.R.; Fenical, W.; Kondratyuk, T.P.; Park, E.-J.; Pezzuto, J.M. Arenamides A–C, cytotoxic NFkappaB inhibitors from the marine actinomycete Salinispora arenicola. J. Nat. Prod. 2009, 72, 396–402. [Google Scholar] [CrossRef]
- Hardy, P.M.; Prout, R.A.; Rydon, H.N. Polypeptides. Part XXV. Synthesis of isariin. J. Chem. Soc. Perkin Trans. 1 1974, 802–808. [Google Scholar] [CrossRef]
- Chandrasekhar, S.; Pavankumarreddy, G.; Sathish, K. Total synthesis of arenamide A and its diastereomer. Tetrahedron Letts. 2009, 50, 6851–6854. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, L.J. ORTEP-3 for Windows—A version of ORTEP-III with a Graphical User Interface (GUI). J. Appl. Crystallogr. 1997, 30, 565. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX suite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]









| (3) | (4) | (5) | ||||
|---|---|---|---|---|---|---|
| Position | δH, mult (J in Hz) | δC | δH, mult (J in Hz) | δC | δH, mult (J in Hz) | δC |
| l-Phe1 | l-Val1 | l-Ala1 | ||||
| 1 | --- | 170.8 | --- | 170.7 | --- | 171.7 g |
| 2 | 4.38, q (7.1) | 54.2 | 4.15, dd (7.8, 5.3) | 57.3 | 4.13, dq (7.4, 6.0) | 48.1 |
| 3 | a 3.01, dd (13.9, 6.1) b 2.93, d (13.9, 8.7) | 36.7 | 2.08, m | 29.7 | 1.28, d (7.4) | 16.7 |
| 4 | --- | 137.2 | 0.87, d (6.8) | 19.0 | ||
| 5/9 | 7.25, m | 129.0 | 0.85a, d (7.0) | 17.4 | ||
| 6/8 | 7.28, m | 128.2 | ||||
| 7 | 7.20, m | 126.5 | ||||
| NH | 7.96, d (7.1) | --- | 7.41, d (7.1) | --- | 7.80, d (6.0) | --- |
| l-Ala2 | l-Ala2 | l-Ala2 | ||||
| 1 | --- | 171.7f | --- | 171.8 | --- | 171.5 |
| 2 | 4.16, dq (7.4, 7.1) | 47.6 | 4.19, dq (8.0, 7.1) | 47.9 | 4.18, dq (7.1, 6.5) | 47.5 |
| 3 | 1.15, d (7.1) | 17.6 | 1.21, d (7.1) | 17.5 | 1.19, d (7.1) | 17.4 |
| NH | 7.84, d (7.4) | --- | 8.03, d (8.0) | --- | 7.89, d (6.5) | --- |
| d-Leu3 | d-Leu3 | d-Leu3 | ||||
| 1 | --- | 171.0 | --- | 171.3 | --- | 171.1 |
| 2 | 4.03, m | 52.0 | 4.04, dd (10.1, 6.5) | 51.9 | 4.03, dd (11.4, 6.3) | 51.8 |
| 3 | 1.46, m | 38.7 | 1.48, m | 38.7 | a 1.49, m b 1.47 a, m | 38.6 |
| 4 | 1.63, m | 24.1 | 1.63, m | 24.1 | 1.63, m | 24.1 |
| 5 | 0.88, d (6.5) | 22.9 | 0.89 b, d (6.7) | 22.9 | 0.89, d (6.7) | 23.0 |
| 6 | 0.81, d (6.6) | 21.0 | 0.81, d (6.6) | 21.0 | 0.81, d (6.5) | 21.1 |
| NH | 8.62, d (6.1) | --- | 8.63, d (6.5) | --- | 8.64, d (6.3) | --- |
| l-Val4 | l-Val4 | l-Val4 | ||||
| 1 | --- | 171.7f | --- | 171.6 | --- | 171.6 g |
| 2 | 4.10, dd (8.6, 7.6) | 58.3 | 4.06 c, m | 58.6 | 4.11, dd (8.2, 6.6) | 58.1 |
| 3 | 1.88, m | 29.8 | 1.86, m | 29.5 | 1.85, m | 29.9 |
| 4 | 0.87, d (6.4) | 18.9 | 0.88 b, d (6.8) | 19.0 | 0.87, d (6.9) | 18.8 |
| 5 | 0.83, d (6.6) | 18.7 | 0.83 d, d (6.8) | 18.7 | 0.83, d (6.7) | 18.7 |
| NH | 7.92 a, d (7.6) | --- | 8.06, d (7.8) | --- | 7.87, d (6.6) | --- |
| Gly5 | Gly5 | Gly5 | ||||
| 1 | --- | 168.9 | --- | 169.0 | --- | 168.8 |
| 2 | a 4.07, dd (16.7, 6.6) b 3.41, dd (16.7, 3.9) | 42.3 | a 4.06 c, m b 3.44, dd (17.1, 3.8) | 42.0 | a 4.06, dd (16.5, 6.6) b 3.43, dd (16.5, 4.0) | 42.3 |
| NH | 7.91 a, dd (6.5, 3.9) | --- | 7.90, dd (5.8, 3.8) | --- | 7.82, dd (6.1, 4.0) | --- |
| HDMLA | HDMLA | HDMLA | ||||
| 1′ | --- | 169.8 | --- | 170.0 | --- | 169.8 |
| 2′ | a 2.51, dd (15.2, 9.8) b 2.25, d (15.2, 1.4) | 37.7 | a 2.52, dd (14.5, 9.6) b 2.23, dd (14.5, 1.4) | 38.0 | a 2.53, dd (14.7, 10.1) b 2.24, dd (14.7, 1.8) | 37.7 |
| 3′ | 4.94, ddd (9.1, 5.1, 1.8) | 74.8 | 4.92, ddd (9.3, 4.8, 1.4) | 75.2 | 4.97, ddd (10.1, 4.8, 1.8) | 74.2 |
| 4′ | 1.68, m | 33.7 | 1.79, m | 33.9 | 1.75, m | 33.8 |
| 5′ | a 1.18 b, m b 0.75, m | 39.5h | a 1.29, m b 0.87, m | 39.6h | a 1.26, m b 0.84, m | 39.6 h |
| 6′ | 1.41, m | 29.3 | 1.44, m | 29.3 | 1.46 a, m | 29.2 |
| 7′ | a 1.19 b, m b 0.95, m | 35.7 | a 1.22 e, m b 0.98, m | 36.0 | a 1.23 b, m b 0.99, m | 35.8 |
| 8′ | 1.20 b, m | 26.1 | a 1.26, m b 1.16, m | 26.1 | a 1.24, m b 1.16, m | 26.1 |
| 9′ | 1.22 c, m | 29.1 | a 1.22 e, m b 1.19, m | 29.1 | 1.23 b, m | 29.0 |
| 10′ | 1.22 c, m | 31.3 | 1.23 e, m | 31.3 | 1.23 b, m | 31.3 |
| 11′ | 1.24, m | 22.1 | 1.25, m | 22.1 | 1.26, m | 22.1 |
| 12′ | 0.84, t (6.9) | 13.9 | 0.85 a, t (7.9) | 13.9 | 0.85, t (7.1) | 13.9 |
| 4′-Me | 0.70, d (6.7) | 14.8 | 0.84 d, d (8.0) | 15.1 | 0.82, d (6.4) | 15.0 |
| 6′-Me | 0.80, d (6.6) | 20.2 | 0.83 d, d (7.4) | 20.0 | 0.83, d (6.7) | 20.2 |
| (6) | (8) | |||
|---|---|---|---|---|
| Position | δH, mult (J in Hz) | δC | δH, mult (J in Hz) | δC |
| l-Phe1 | l-Phe1 | |||
| 1 | --- | 170.9 | --- | 170.8 |
| 2 | 4.38, m | 54.3 | 4.34, q (7.4) | 54.5 |
| 3 | a 2.97, dd (13.9, 6.1) b 2.95, dd (13.9, 8.4) | 36.7 | 2.98, m | 36.7 |
| 4 | --- | 137.1 | --- | 137.2 |
| 5/9 | 7.26, m | 128.9 | 7.26, m | 129.1 |
| 6/8 | 7.28, m | 128.3 | 7.26, m | 128.2 |
| 7 | 7.21, m | 126.6 | 7.20, m | 126.5 |
| NH | 8.14, d (6.5) | --- | 8.03, d (6.7) | --- |
| l-Abu2 | l-Ala2 | |||
| 1 | --- | 171.0 | --- | 171.8 |
| 2 | 4.08 a, m | 52.9 | 4.19, dq (8.1, 7.1) | 47.6 |
| 3 | a 1.74, m b 1.48 b, m | 24.4 | 1.15, d (7.1) | 17.6 |
| 4 | 0.73, t (7.2) | 9.5 | ||
| NH | 7.67, d (8.0) | --- | 7.83, d (8.1) | --- |
| d-Leu3 | d-Leu3 | |||
| 1 | --- | 171.1 | --- | 171.0 |
| 2 | 4.09 a, m | 51.8 | 4.03, m | 52.0 |
| 3 | 1.47 b, m | 38.6 | a 1.47, m b 1.44, m | 38.7 |
| 4 | 1.61, m | 24.1 | 1.63, m | 24.1 |
| 5 | 0.88, d (6.5) | 22.8 | 0.89, d (6.5) | 22.9 |
| 6 | 0.81, d (6.8) | 21.2 | 0.82 d, d (6.5) | 21.1 |
| NH | 8.62, d (6.3) | --- | 8.63, d (6.0) | --- |
| l-Val4 | l-Val4 | |||
| 1 | --- | 171.6 | --- | 171.7 |
| 2 | 4.15, dd (8.9, 8.7) | 58.1 | 4.10, dd (8.6, 7.9) | 58.3 |
| 3 | 1.88, m | 29.9 | 1.87, m | 29.8 |
| 4 | 0.86, d (6.7) | 18.8 | 0.88 c, d (6.2) | 18.9 |
| 5 | 0.83 f, d (6.6) | 18.7 | 0.81 d, d (6.1) | 18.7 |
| NH | 7.86, d (8.9) | --- | 7.88, d (7.9) | --- |
| Gly5 | Gly5 | |||
| 1 | --- | 168.6 | --- | 168.9 |
| 2 | a 4.01, dd (16.7, 6.3) b 3.42, dd (16.7, 4.7) | 42.4 | a 4.07, dd (16.6, 6.4) b 3.40, dd (16.6, 4.1) | 42.3 |
| NH | 7.85, dd (5.6, 4.7) | --- | 7.93, dd (6.4, 4.1) | --- |
| HDMLA | HMLA | |||
| 1′ | --- | 169.8 | --- | 169.8 |
| 2′ | a 2.53, dd (14.8, 9.4) b 2.24, dd (14.8, 1.2) | 37.9 | a 2.48, dd (14.9, 9.2) b 2.24, dd (14.9, 1.8) | 37.0 |
| 3′ | 4.94, ddd (9.1, 3.2, 1.8) | 74.7 | 4.90, ddd (9.2, 5.7, 1.8) | 75.1 |
| 4′ | 1.66, m | 33.9 | 1.50, m | 36.1 |
| 5’ | a 1.17 e, m b 0.75, m | 39.6f | a 1.19 b, m b 0.87 c, m | 31.4 |
| 6′ | 1.41, m | 29.2 | a 1.19 b, m b 1.09, m | 26.5 |
| 7′ | a 1.19, m b 0.95, m | 35.8 | 1.18 b, m | 29.3 |
| 8′ | a 1.25 c, m b 1.17 e, m | 26.1 | 1.23 a, m | 28.9 |
| 9′ | 1.22 d, m | 29.1 | 1.23 a, m | 28.7 |
| 10′ | 1.22 d, m | 31.3 | 1.23 a, m | 31.3 |
| 11′ | 1.24 c, m | 22.1 | 1.25, m | 22.1 |
| 12′ | 0.83 f, t (6.9) | 13.9 | 0.85, t (6.6) | 13.9 |
| 4′-Me | 0.69, d (6.7) | 14.8 | 0.67, d (6.9) | 14.5 |
| 6′-Me | 0.80, d (6.8) | 20.1 | ||

| Compound | aa1 | aa2 | aa3 | aa4 | aa5 | Compound | aa1 | aa2 | aa3 | aa4 | aa5 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| scopularide A [2] | l-Phe | l-Ala | d-Leu | l-Val | Gly | iso-isariin D [17] | l-Ala | l-Ala | d-Leu | l-Val | Gly |
| scopularide B [2] | l-Phe | l-Ala | d-Leu | l-Val | Gly | isariin G2 [15] | l-Ala | l-Ala | d-Leu | l-Val | Gly |
| scopularide C | l-Phe | l-Ala | d-Leu | l-Val | Gly | scopularide E | l-Ala | l-Ala | d-Leu | l-Val | Gly |
| scopularide H | l-Phe | l-Ala | d-Leu | l-Val | Gly | isariin F1 [15] | Abu/Aib | l-Ala | d-Leu | l-Val | Gly |
| chrysogeamide A [6] | l-Val | l-Ala | d-Leu | l-Val | Gly | scopularide F | l-Phe | l-Abu | d-Leu | l-Val | Gly |
| chrysogeamide B [6] | l-Val | l-Ala | d-Leu | l-Val | Gly | chrysogeamide C [6] | l-Phe | l-Ala | d-Leu | d-Leu | Gly |
| scopularide D | l-Val | l-Ala | d-Leu | l-Val | Gly | chrysogeamide D [6] | l-Phe | l-Ala | d-Leu | d-Leu | Gly |
| nodupetide [9] | l-Val | l-Ala | d-Leu | l-Val | Gly | chrysogeamide E [6] | l-Phe | l-Ala | d-Leu | d-Leu | Gly |
| isariin A [10,11] | l-Val | l-Ala | d-Leu | l-Val | Gly | chrysogeamide F [6] | l-Phe | l-Ala | d-Leu | l-Pro | Gly |
| isariin B [12,13,14] | l-Val | l-Ala | d-Leu | l-Val | Gly | chrysogeamide G [6] | l-Phe | l-Ala | d-Leu | l-Pro | Gly |
| iso-isariin B [16] | l-Val | l-Ala | d-Leu | l-Val | Gly | scopularide G | l-Val | l-Abu | d-Leu | l-Val | Gly |
| isariin C2 [15] | l-Val | l-Ala | d-Leu | l-Val | Gly | emericellamide A [7] | l-Ala | l-Ala | l-Leu | l-Val | Gly |
| isariin E [15] | l-Val | l-Ala | d-Leu | l-Val | Gly | emericellamide B [7] | l-Ala | l-Ala | l-Leu | l-Val | Gly |
| isariin F2 [15] | l-Val | l-Ala | d-Leu | l-Val | Gly | emericellamide C [8] | l-Ala | l-Ala | l-Leu | l-Val | Gly |
| isariin G1 [15] | l-Val | l-Ala | d-Leu | l-Val | Gly | emericellamide D [8] | l-Ala | l-Ala | l-Leu | l-Val | Gly |
| oryzamide A [5] | l-Leu | l-Ala | d-Leu | l-Val | Gly | emericellamide E [8] | l-Ala | l-Ala | l-Leu | l-Val | Gly |
| oryzamide B [5] | l-Tyr | l-Ala | d-Leu | l-Val | Gly | emericellamide F [8] | l-Ala | l-Ala | l-Leu | l-Val | Gly |
| oryzamide C [5] | l-Met | l-Ala | d-Leu | l-Val | Gly | arenamide A [18] | l-Phe | l-Ala | l-Leu | l-Val | Gly |
| isariin C [13,14] | l-Ala | l-Ala | d-Leu | l-Val | Gly | arenamide B [18] | l-Phe | l-Ala | l-Leu | l-Val | Gly |
| isariin D [13,14] | l-Ala | l-Ala | d-Leu | l-Val | Gly | arenamide C [18] | l-Met | l-Ala | l-Leu | l-Val | Gly |

| Structure a | |||||||||
| compound | n | R | compound | n | R | ||||
| isariin E * [15] | 1 | H | iso-isariin B * [16] | 3 | Me | ||||
| iso-isariin D [17] | 1 | Me | isariin B [12,13,14] | 4 | H | ||||
| nodupetide [9] | 1 | Me | isariin C * [13,14] | 4 | H | ||||
| chrysogeamide D [6] | 1 | Me | isariin F1* [15] | 4 | H | ||||
| isariin C2 * [15] | 2 | H | isariin G1 * [15] | 5 | H | ||||
| isariin D * [13,14] | 2 | H | isariin A [10,11,19] | 7 | H | ||||
| isariin F2 * [15] | 3 | H | isariin G2 * [15] | 7 | H | ||||
| Structure b | |||||||||
| compound | n | R1 | R2 | R3 | compound | n | R1 | R2 | R3 |
| scopularide B [2] | 1 | H | Me | H | arenamide A [18,20] | 3 | H | Me | H |
| chrysogeamide A [6] | 1 | H | Me | H | arenamide C [18,20] | 3 | H | Me | H |
| chrysogeamide E [6] | 1 | H | Me | H | emericellamide A [7] | 3 | Me | Me | H |
| chrysogeamide F [6] | 1 | H | Me | H | emericellamide C [8] | 3 | Me | H | H |
| arenamide B [18,20] | 1 | H | Me | H | emericellamide E [8] | 5 | Me | H | H |
| scopularide A [2] | 3 | H | Me | H | emericellamide F [8] | 5 | H | Me | H |
| chrysogeamide B [6] | 3 | H | Me | H | scopularide H | 5 | H | Me | H |
| chrysogeamide C [6] | 3 | H | Me | H | scopularide C | 5 | H | Me | Me |
| chrysogeamide G [6] | 3 | H | Me | H | scopularide D | 5 | H | Me | Me |
| oryzamide A [5] | 3 | H | Me | H | scopularide E | 5 | H | Me | Me |
| oryzamide B [5] | 3 | H | Me | H | scopularide F | 5 | H | Me | Me |
| oryzamide C [5] | 3 | H | Me | H | scopularide G | 5 | H | Me | Me |
| emericellamide D [8] | 3 | H | Me | H | emericellamide B [7] | 5 | Me | Me | Me |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elbanna, A.H.; Khalil, Z.G.; Bernhardt, P.V.; Capon, R.J. Scopularides Revisited: Molecular Networking Guided Exploration of Lipodepsipeptides in Australian Marine Fish Gastrointestinal Tract-Derived Fungi. Mar. Drugs 2019, 17, 475. https://doi.org/10.3390/md17080475
Elbanna AH, Khalil ZG, Bernhardt PV, Capon RJ. Scopularides Revisited: Molecular Networking Guided Exploration of Lipodepsipeptides in Australian Marine Fish Gastrointestinal Tract-Derived Fungi. Marine Drugs. 2019; 17(8):475. https://doi.org/10.3390/md17080475
Chicago/Turabian StyleElbanna, Ahmed H., Zeinab G. Khalil, Paul V. Bernhardt, and Robert J. Capon. 2019. "Scopularides Revisited: Molecular Networking Guided Exploration of Lipodepsipeptides in Australian Marine Fish Gastrointestinal Tract-Derived Fungi" Marine Drugs 17, no. 8: 475. https://doi.org/10.3390/md17080475
APA StyleElbanna, A. H., Khalil, Z. G., Bernhardt, P. V., & Capon, R. J. (2019). Scopularides Revisited: Molecular Networking Guided Exploration of Lipodepsipeptides in Australian Marine Fish Gastrointestinal Tract-Derived Fungi. Marine Drugs, 17(8), 475. https://doi.org/10.3390/md17080475