Discovery of Bioactive Indole-Diketopiperazines from the Marine-Derived Fungus Penicillium brasilianum Aided by Genomic Information
Abstract
:1. Introduction
2. Results and Discussion
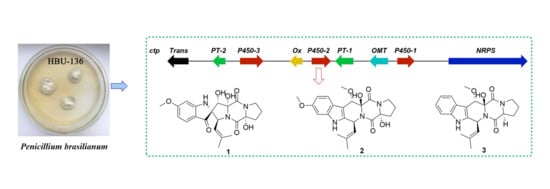
2.1. Identification and Analysis of the Indole-Diketopiperazine BGC
2.2. Structure Elucidation
2.3. Biological Activities Screening
3. Materials and Methods
3.1. Instrumentation
3.2. Genome Sequencing and Bioinformatics Analysis
3.3. Fungal Material
3.4. Fermentation and Purification
3.5. Biological Assay
3.6. ECD Spectrum Measurement and Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piel, J.; Hoang, K.; Moore, B.S. Natural metabolic diversity encoded by the enterocin biosynthesis gene cluster. J. Am. Chem. Soc. 2000, 122, 5415–5416. [Google Scholar] [CrossRef]
- Xu, W.; Dhingra, S.; Chooi, Y.H.; Calvo, A.M.; Lin, H.C.; Tang, Y. The fumagillin biosynthetic gene cluster in Aspergillus fumigatus encodes a cryptic terpene cyclase involved in the formation of β-trans-bergamotene. J. Am. Chem. Soc. 2013, 135, 4616–4619. [Google Scholar]
- Wang, J.F.; He, W.J.; Huang, X.L.; Tian, X.P.; Liao, S.R.; Yang, B.; Wang, F.Z.; Zhou, X.J.; Liu, Y.H. Antifungal new oxepine-containing alkaloids and xanthones from the deep-sea-derived fungus Aspergillus versicolor SCSIO 05879. J. Agric. Food Chem. 2016, 64, 2910–2916. [Google Scholar] [CrossRef] [PubMed]
- Song, F.H.; Liu, X.R.; Guo, H.; Ren, B.; Chen, C.X.; Piggott, A.M.; Yu, K.; Gao, H.; Wang, Q.; Liu, M.; et al. Brevianamides with antitubercular potential from a marine-derived isolate of Aspergillus versicolor. Org. Lett. 2012, 14, 4770–4773. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Z.; Sun, M.W.; Hao, H.L.; Lu, C.H. Avertoxins A–D, Prenyl asteltoxin derivatives from Aspergillus versicolor Y10, an endophytic fungus of Huperzia serrate. J. Nat. Prod. 2015, 78, 3067–3070. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Miao, M.M.; Du, G.; Li, X.N.; Shang, S.Z.; Zhao, W.; Liu, Z.H.; Yang, G.Y.; Che, C.T.; Hu, Q.F.; et al. Aspergillines A–E, highly oxygenated hexacyclic indole–tetrahydrofuran–tetramic acid derivatives from Aspergillus versicolor. Org. Lett. 2014, 16, 5016–5019. [Google Scholar] [CrossRef]
- Son, S.; Hong, Y.S.; Jang, M.; Heo, K.T.; Lee, B.; Jang, J.P.; Kim, J.W.; Ryoo, I.J.; Kim, W.G.; Ko, S.K.; et al. Genomics-driven discovery of chlorinated cyclic hexapeptides ulleungmycins A and B from a Streptomyces species. J. Nat. Prod. 2017, 80, 3025–3031. [Google Scholar] [CrossRef]
- Hu, Y.Y.; Wang, M.; Wu, C.Y.; Tan, Y.; Li, J.; Hao, X.M.; Duan, Y.B.; Guan, Y.; Shang, X.Y.; Wang, Y.G.; et al. Identification and proposed relative and absolute configurations of niphimycins C–E from the marine-derived Streptomyces sp. IMB7-145 by genomic analysis. J. Nat. Prod. 2018, 81, 178–187. [Google Scholar] [CrossRef]
- Xu, X.K.; Zhou, H.B.; Liu, Y.; Liu, X.T.; Fu, J.; Li, A.Y.; Li, Y.Z.; Shen, Y.M.; Bian, X.Y.; Zhang, Y.M. Heterologous Expression Guides Identification of the Biosynthetic Gene Cluster of Chuangxinmycin, an Indole Alkaloid Antibiotic. J. Nat. Prod. 2018, 81, 1060–1064. [Google Scholar] [CrossRef]
- Zhu, A.; Zhang, X.W.; Zhang, M.; Li, W.; Ma, Z.Y.; Zhu, H.J.; Cao, F. Aspergixanthones I–K, new anti-Vibrio prenylxanthones from the marine-derived fungus Aspergillus sp. ZA-01. Mar. Drugs 2018, 16, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.K.; Zhang, B.; Gao, T.; Yang, M.Y.; Zhao, G.Z.; Zhu, H.J.; Cao, F. A pair of enantiomeric 5-oxabicyclic [4.3.0] lactam derivatives and one new polyketide from the marine-derived fungus Penicillium griseofulvum. Nat. Prod. Res. 2018, 32, 2366–2369. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Meng, Z.H.; Mu, X.; Yue, Y.F.; Zhu, H.J. Absolute configuration of bioactive azaphilones from the marine-derived fungus Pleosporales sp. CF09-1. J. Nat. Prod. 2019, 82, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Yang, M.Y.; Zhang, Y.H.; Shao, C.L.; Wang, C.Y.; Hu, L.D.; Cao, F.; Zhu, H.J. Absolute configurations of 14, 15-hydroxylated prenylxanthones from a marine-derived Aspergillus sp. fungus by chiroptical methods. Sci. Rep. 2018, 8, 10621–10630. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.Z.; Fang, Y.C.; Zhu, T.J.; Zhang, M.; Lin, A.Q.; Gu, Q.Q.; Zhu, W.M. Seven new prenylated indole diketopiperazine alkaloids from holothurian-derived fungus Aspergillus fumigatus. Tetrahedron 2008, 64, 7986–7991. [Google Scholar] [CrossRef]
- Cui, C.B.; Kakeya, H.; Osada, H. Novel mammalian cell cycle inhibitors, spirotryprostatins A and B, produced by Aspergillus fumigatus, which inhibit mammalian cell cycle at G2/M phase. Tetrahedron 1996, 52, 12651–12666. [Google Scholar] [CrossRef]
- Kato, N.; Suzuki, H.; Takagi, H.; Asami, Y.; Kakeya, H.; Uramoto, M.; Usui, T.; Takahashi, S.; Sugimoto, Y.; Osada, H. Identification of cytochrome P450s required for fumitremorgin biosynthesis in Aspergillus fumigatus. ChemBioChem. 2010, 10, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Maiya, S.; Grundmann, A.; Li, S.M.; Turner, G. The fumitremorgin gene cluster of Aspergillus fumigatus: Identification of a gene encoding brevianamide F synthetase. ChemBioChem. 2010, 7, 1062–1069. [Google Scholar] [CrossRef]
- Grundmann, A.; Kuznetsova, T.; Afiyatullov, S.S.; Li, S.M. FtmPT2, an N-Prenyltransferase from Aspergillus fumigatus, catalyses the last step in the biosynthesis of fumitremorgin B. ChemBioChem 2008, 9, 2059–2063. [Google Scholar] [CrossRef]
- Tsunematsu, Y.; Ishiuchi, K.; Hotta, K.; Watanabe, K. Yeast-based genome mining, production and mechanistic studies of the biosynthesis of fungal polyketide and peptide natural products. Nat. Prod. Rep. 2013, 30, 1139–1149. [Google Scholar] [CrossRef]
- Gu, B.; He, S.; Yan, X.; Zhang, L. Tentative biosynthetic pathways of some microbial diketopiperazines. Appl. Microbiol. Biot. 2013, 97, 8439–8453. [Google Scholar] [CrossRef] [PubMed]
- Tsunematsu, Y.; Ishikawa, N.; Wakana, D.; Goda, Y.; Noguchi, H.; Moriya, H.; Hotta, K.; Watanabe, K. Distinct mechanisms for spiro-carbon formation reveal biosynthetic pathway crosstalk. Nat. Chem. Biol. 2013, 9, 818–827. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.W.; Li, S.Y. Expansion of chemical space for natural products by uncommon P450 reactions. Nat. Prod. Rep. 2017, 34, 1061–1089. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Li, D.Y.; Li, Y.C.; Hua, H.M.; Ma, E.L.; Li, Z.L. Caryophyllene sesquiterpenes from the marine-derived fungus Ascotricha sp. ZJ-M-5 by the one strain–many compounds strategy. J. Nat. Prod. 2014, 77, 1367–1371. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.L.; Bai, J.; Zhang, L.M.; Wu, X.; Zhang, L.; Pei, Y.H.; Jing, Y.K.; Hua, H.M. 2,5-Diketopiperazines from the Marine-Derived Fungus Aspergillus fumigatus YK-7. Chem. Biodivers. 2012, 9, 385–393. [Google Scholar] [CrossRef]
- Yu, L.Y.; Ding, W.J.; Wang, Q.Q.; Ma, Z.J.; Xu, X.W.; Zhao, X.F.; Chen, Z. Induction of cryptic bioactive 2, 5-diketopiperazines in fungus Penicillium sp. DT-F29 by microbial co-culture. Tetrahedron 2017, 73, 907–914. [Google Scholar] [CrossRef]
- Xue, H.; Lu, C.H.; Liang, L.Y.; Shen, Y.M. Secondary Metabolites of Aspergillus sp. CM9a, an Endophytic Fungus of Cephalotaxus mannii. Rec. Nat. Prod. 2012, 6, 28–34. [Google Scholar]
- Wang, F.Q.; Tong, Q.Y.; Ma, H.R.; Xu, H.F.; Hu, S.; Ma, W.; Xue, Y.B.; Liu, J.J.; Wang, J.P.; Song, H.P.; et al. Indole diketopiperazines from endophytic Chaetomium sp. 88194 induce breast cancer cell apoptotic death. Sci. Rep. 2015, 5, 9294. [Google Scholar] [CrossRef]
- Meng, L.H.; Du, F.Y.; Li, X.M.; Pedpradab, P.; Xu, G.M.; Wang, B.G. Rubrumazines A–C, indolediketopiperazines of the isoechinulin class from Eurotium rubrum MA-150, a fungus obtained from marine mangrove-derived rhizospheric soil. J. Nat. Prod. 2015, 78, 909–913. [Google Scholar] [CrossRef]
- Scudiere, D.A.; Shoemaker, R.H.; Paull, K.D.; Monks, A.; Tierney, S.; Nofziger, T.H.; Currens, M.J.; Seniff, D.; Boyd, M.R. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 1988, 48, 4827–4833. [Google Scholar]
- Appendino, G.; Gibbons, S.; Giana, A.; Pagani, A.; Grassi, G.; Stavri, M.; Smith, E.; Rahman, M.M. Antibacterial cannabinoids from Cannabis sativa: A structure−Activity study. J. Nat. Prod. 2008, 71, 1427–1430. [Google Scholar] [CrossRef] [PubMed]
- Bruhn, T.; Schaumlöffel, A.; Hemberger, Y.; Bringmann, G. SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef] [PubMed]






| No. | 1 | 2 | 3 |
|---|---|---|---|
| 1 | 7.86, s | 7.97, s | |
| 4 | 7.50, d (8.7) | 7.48, d (8.4) | 7.58, d (7.6) |
| 5 | 6.45, dd (8.7, 2.0) | 6.80, dd (8.4, 2.0) | 7.17, dtd (8.0, 7.6, 0.9) |
| 6 | 7.17, dtd (8.0, 7.6, 0.9) | ||
| 7 | 6.27, d (2.0) | 6.86, d (2.0) | 7.37, d (7.6) |
| 8 | 4.70, s | 5.02, s | 4.79, s |
| 12 | 4.38, dd (10.7, 6.3) | ||
| 13 | 2.35, m | 2.40, m | 2.50, m |
| 2.27, m | 1.98, m | 2.01, m | |
| 14 | 2.15, m | 2.10, s | 2.12, m |
| 2.00, m | 1.98, m | 2.01, m | |
| 15 | 3.74, m | 3.80, m | 3.76, m |
| 3.54, m | 3.70, m | 3.71, m | |
| 18 | 4.83, d (9.5) | 6.36, d (9.6) | 6.68, d (9.7) |
| 19 | 4.79, d (9.5) | 5.33, d (9.6) | 5.57, d (9.7) |
| 21 | 1.56, s | 2.04, s | 1.80, s |
| 22 | 1.79, s | 1.77, s | 2.07, s |
| 8-OH | 4.96, brs | ||
| 12-OH | 5.14, brs | ||
| 6-OCH3 | 3.85, s | 3.82, s | |
| 9-OH | 8.51, brs | ||
| 8-OCH3 | 3.30, s | 3.38, s |
| No. | 1 | 2 | 3 |
|---|---|---|---|
| 2 | 75.0, C | 132.7, C | 135.2, C |
| 3 | 200.6, C | 104.8, C | 105.7, C |
| 3a | 112.4, C | 122.5, C | 128.5, C |
| 4 | 127.4, CH | 118.9, CH | 118.2, CH |
| 5 | 110.8, CH | 110.1, CH | 120.7, CH |
| 6 | 169.7, C | 156.6, C | 122.3, CH |
| 7 | 94.9, CH | 95.3, CH | 111.3, CH |
| 7a | 165.4, C | 137.1, C | 135.9, C |
| 8 | 73.9, CH | 74.9, CH | 76.8, CH |
| 9 | 86.1, C | 85.7, C | 84.9, C |
| 11 | 167.4, C | 166.3, C | 167.2, C |
| 12 | 90.2, C | 86.7, C | 60.1, CH |
| 13 | 35.1, CH2 | 36.5, CH2 | 29.8, CH2 |
| 14 | 21.3, CH2 | 19.2, CH2 | 22.3, CH2 |
| 15 | 45.3, CH2 | 45.4, CH2 | 46.0, CH2 |
| 17 | 166.0, C | 165.9, C | 166.0, C |
| 18 | 55.7, CH | 49.3, CH | 49.2, CH |
| 19 | 119.4, CH | 122.8, CH | 123.6, CH |
| 20 | 142.4, C | 138.2, C | 138.2, C |
| 21 | 26.1, CH3 | 18.4, CH3 | 26.2, CH3 |
| 22 | 18.8, CH3 | 26.1, CH3 | 18.4, CH3 |
| 6-OCH3 | 56.0, CH3 | 55.8, OCH3 | |
| 8-OCH3 | 57.0, OCH3 | 56.8, OCH3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.-H.; Geng, C.; Zhang, X.-W.; Zhu, H.-J.; Shao, C.-L.; Cao, F.; Wang, C.-Y. Discovery of Bioactive Indole-Diketopiperazines from the Marine-Derived Fungus Penicillium brasilianum Aided by Genomic Information. Mar. Drugs 2019, 17, 514. https://doi.org/10.3390/md17090514
Zhang Y-H, Geng C, Zhang X-W, Zhu H-J, Shao C-L, Cao F, Wang C-Y. Discovery of Bioactive Indole-Diketopiperazines from the Marine-Derived Fungus Penicillium brasilianum Aided by Genomic Information. Marine Drugs. 2019; 17(9):514. https://doi.org/10.3390/md17090514
Chicago/Turabian StyleZhang, Ya-Hui, Ce Geng, Xing-Wang Zhang, Hua-Jie Zhu, Chang-Lun Shao, Fei Cao, and Chang-Yun Wang. 2019. "Discovery of Bioactive Indole-Diketopiperazines from the Marine-Derived Fungus Penicillium brasilianum Aided by Genomic Information" Marine Drugs 17, no. 9: 514. https://doi.org/10.3390/md17090514
APA StyleZhang, Y.-H., Geng, C., Zhang, X.-W., Zhu, H.-J., Shao, C.-L., Cao, F., & Wang, C.-Y. (2019). Discovery of Bioactive Indole-Diketopiperazines from the Marine-Derived Fungus Penicillium brasilianum Aided by Genomic Information. Marine Drugs, 17(9), 514. https://doi.org/10.3390/md17090514